[English] 日本語
 Yorodumi
Yorodumi- EMDB-4934: Cryo-EM structure of the microsporidian ribosome: State 1, Multib... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4934 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the microsporidian ribosome: State 1, Multibody-refined map body 3 (SSU-head) | |||||||||
 Map data Map data | Cryo-EM structure of the microsporidian ribosome: State 1, Multibody-refined map body 3 (SSU-head) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Vairimorpha necatrix (fungus) Vairimorpha necatrix (fungus) | |||||||||
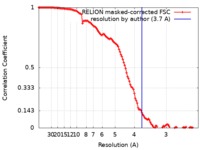
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Barandun J / Hunziker M / Vossbrinck CR / Klinge S | |||||||||
| Funding support |  United States, United States,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. Authors: Jonas Barandun / Mirjam Hunziker / Charles R Vossbrinck / Sebastian Klinge /   Abstract: Microsporidia are eukaryotic parasites that infect essentially all animal species, including many of agricultural importance, and are significant opportunistic parasites of humans. They are ...Microsporidia are eukaryotic parasites that infect essentially all animal species, including many of agricultural importance, and are significant opportunistic parasites of humans. They are characterized by having a specialized infection apparatus, an obligate intracellular lifestyle, rudimentary mitochondria and the smallest known eukaryotic genomes. Extreme genome compaction led to minimal gene sizes affecting even conserved ancient complexes such as the ribosome. In the present study, the cryo-electron microscopy structure of the ribosome from the microsporidium Vairimorpha necatrix is presented, which illustrates how genome compaction has resulted in the smallest known eukaryotic cytoplasmic ribosome. Selection pressure led to the loss of two ribosomal proteins and removal of essentially all eukaryote-specific ribosomal RNA (rRNA) expansion segments, reducing the rRNA to a functionally conserved core. The structure highlights how one microsporidia-specific and several repurposed existing ribosomal proteins compensate for the extensive rRNA reduction. The microsporidian ribosome is kept in an inactive state by two previously uncharacterized dormancy factors that specifically target the functionally important E-site, P-site and polypeptide exit tunnel. The present study illustrates the distinct effects of evolutionary pressure on RNA and protein-coding genes, provides a mechanism for ribosome inhibition and can serve as a structural basis for the development of inhibitors against microsporidian parasites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4934.map.gz emd_4934.map.gz | 137.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4934-v30.xml emd-4934-v30.xml emd-4934.xml emd-4934.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4934_fsc.xml emd_4934_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4934.png emd_4934.png | 209.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4934 http://ftp.pdbj.org/pub/emdb/structures/EMD-4934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4934 | HTTPS FTP |
-Related structure data
| Related structure data |  4931C  4932C  4933C  4935C  6rm3C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11075 (Title: Single-particle cryo-EM dataset of the Vairimorpha necatrix ribosome EMPIAR-11075 (Title: Single-particle cryo-EM dataset of the Vairimorpha necatrix ribosomeData size: 1.0 TB Data #1: Unaligned multi-frame micrographs of the Vairimorpha necatrix ribosome [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4934.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4934.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the microsporidian ribosome: State 1, Multibody-refined map body 3 (SSU-head) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
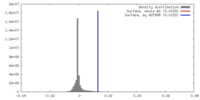
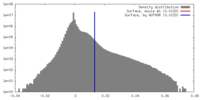
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Microsporidian Ribosome
| Entire | Name: Microsporidian Ribosome |
|---|---|
| Components |
|
-Supramolecule #1: Microsporidian Ribosome
| Supramolecule | Name: Microsporidian Ribosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#78 |
|---|---|
| Source (natural) | Organism:  Vairimorpha necatrix (fungus) Vairimorpha necatrix (fungus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 11 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 2 / Number real images: 3284 / Average exposure time: 0.25 sec. / Average electron dose: 5.55 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus min: 4.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: OTHER |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)