[English] 日本語
 Yorodumi
Yorodumi- EMDB-4874: Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4874 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex I / NADH dehydrogenase / Mitochondrion Proton pumping / Ubiquinone / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationlipoate biosynthetic process / NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / membrane protein complex / NADH dehydrogenase complex / ubiquinone biosynthetic process / mitochondrial [2Fe-2S] assembly complex / oxidoreductase activity, acting on NAD(P)H / iron-sulfur cluster assembly / protein import into mitochondrial matrix ...lipoate biosynthetic process / NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / membrane protein complex / NADH dehydrogenase complex / ubiquinone biosynthetic process / mitochondrial [2Fe-2S] assembly complex / oxidoreductase activity, acting on NAD(P)H / iron-sulfur cluster assembly / protein import into mitochondrial matrix / ubiquinone binding / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / catalytic complex / quinone binding / ATP synthesis coupled electron transport / aerobic respiration / respiratory electron transport chain / electron transport chain / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress / oxidoreductase activity / mitochondrial inner membrane / protein-containing complex binding / mitochondrion / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) | |||||||||
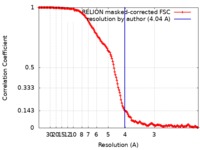
| Method | single particle reconstruction / cryo EM / Resolution: 4.04 Å | |||||||||
 Authors Authors | Parey K / Vonck J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease. Authors: Kristian Parey / Outi Haapanen / Vivek Sharma / Harald Köfeler / Thomas Züllig / Simone Prinz / Karin Siegmund / Ilka Wittig / Deryck J Mills / Janet Vonck / Werner Kühlbrandt / Volker Zickermann /    Abstract: Respiratory complex I is a redox-driven proton pump, accounting for a large part of the electrochemical gradient that powers mitochondrial adenosine triphosphate synthesis. Complex I dysfunction is ...Respiratory complex I is a redox-driven proton pump, accounting for a large part of the electrochemical gradient that powers mitochondrial adenosine triphosphate synthesis. Complex I dysfunction is associated with severe human diseases. Assembly of the one-megadalton complex I in the inner mitochondrial membrane requires assembly factors and chaperones. We have determined the structure of complex I from the aerobic yeast by electron cryo-microscopy at 3.2-Å resolution. A ubiquinone molecule was identified in the access path to the active site. The electron cryo-microscopy structure indicated an unusual lipid-protein arrangement at the junction of membrane and matrix arms that was confirmed by molecular simulations. The structure of a complex I mutant and an assembly intermediate provide detailed molecular insights into the cause of a hereditary complex I-linked disease and complex I assembly in the inner mitochondrial membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4874.map.gz emd_4874.map.gz | 337.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4874-v30.xml emd-4874-v30.xml emd-4874.xml emd-4874.xml | 64.1 KB 64.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4874_fsc.xml emd_4874_fsc.xml | 16.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4874.png emd_4874.png | 135.5 KB | ||
| Filedesc metadata |  emd-4874.cif.gz emd-4874.cif.gz | 14.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4874 http://ftp.pdbj.org/pub/emdb/structures/EMD-4874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4874 | HTTPS FTP |
-Related structure data
| Related structure data |  6rfsMC  4872C  4873C  6rfqC  6rfrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4874.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4874.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Mitochondrial NADH:ubiquinone oxidoreductase
+Supramolecule #1: Mitochondrial NADH:ubiquinone oxidoreductase
+Macromolecule #1: Subunit NUAM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #2: Subunit NUBM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #3: Subunit NUCM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #4: Subunit NIMM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #5: Subunit NUEM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #6: Subunit NUFM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #7: Subunit NUGM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #8: Subunit NUHM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #9: Subunit NUIM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #10: Subunit NUJM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #11: Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #12: Subunit NULM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #13: Subunit NUMM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #14: Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Com...
+Macromolecule #15: Subunit NB4M of protein NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #16: Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Com...
+Macromolecule #17: Subunit NI2M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #18: Subunit NESM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #19: Subunit NUPM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #20: Subunit NB6M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #21: Subunit NUXM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #22: Subunit NUZM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #23: Subunit NIAM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #24: Subunit NEBM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #25: Subunit NB2M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #26: Subunit NIDM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #27: Subunit NUVM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #28: Subunit NI8M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #29: Subunit NI9M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #30: Subunit N7BM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #31: Subunit NUUM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #32: Subunit NB5M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #33: Subunit NUNM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #34: Subunit NU1M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #35: Subunit NU2M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #36: Subunit NU3M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #37: Subunit NU4M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #38: Subunit NU5M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #39: Subunit NU6M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #40: Subunit NB8M of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #41: Subunit NIPM of NADH:Ubiquinone Oxidoreductase (Complex I)
+Macromolecule #42: IRON/SULFUR CLUSTER
+Macromolecule #43: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #44: FLAVIN MONONUCLEOTIDE
+Macromolecule #45: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #46: ZINC ION
+Macromolecule #47: S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 5964 / Average exposure time: 8.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 45872 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -2.0 µm / Nominal magnification: 200000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6rfs: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)