[English] 日本語
 Yorodumi
Yorodumi- EMDB-48234: Structure of zebrafish OTOP1 in nanodisc in the presence of inhib... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of zebrafish OTOP1 in nanodisc in the presence of inhibitor C11 | |||||||||
 Map data Map data | Refinement map for structure of zebrafish OTOP1 in nanodisc in the presence of inhibitor C11 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | proton channel / ion channel / OTOP / Otopetrin / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationotolith formation / otolith mineralization / proton channel activity / inner ear morphogenesis / cholesterol binding / negative regulation of type II interferon-mediated signaling pathway / microvillus / proton transmembrane transport / cellular response to insulin stimulus / identical protein binding ...otolith formation / otolith mineralization / proton channel activity / inner ear morphogenesis / cholesterol binding / negative regulation of type II interferon-mediated signaling pathway / microvillus / proton transmembrane transport / cellular response to insulin stimulus / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Burendei B / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structure-guided discovery of Otopetrin 1 inhibitors reveals druggable binding sites at the intrasubunit interface. Authors: Batuujin Burendei / Joshua P Kaplan / Gerardo M Orellana / Emily R Liman / Stefano Forli / Andrew B Ward /  Abstract: Proton conductance across cell membranes serves many biological functions, ranging from the regulation of intracellular and extracellular pH to the generation of electrical signals that lead to ...Proton conductance across cell membranes serves many biological functions, ranging from the regulation of intracellular and extracellular pH to the generation of electrical signals that lead to sour taste perception. Otopetrins (OTOPs) are a conserved, eukaryotic family of proton-selective ion channels, one of which (OTOP1) serves as a gustatory sensor for sour tastes and ammonium chloride. As the functional properties and structures of OTOP channels were only recently described, there are presently few tools available to modulate their activity. Here, we perform subsequent rounds of molecular docking-based virtual screening against the structure of zebrafish OTOP1, followed by functional testing using whole-cell patch-clamp electrophysiology, and identify several small molecule inhibitors that are effective in the low-to-mid µM range. Cryo-electron microscopy structures reveal inhibitor binding sites in the intrasubunit interface that are validated by functional testing of mutant channels. Our findings reveal pockets that can be targeted for small molecule discovery to develop modulators for Otopetrins. Such modulators can serve as useful toolkit molecules for future investigations of structure-function relationships or physiological roles of Otopetrins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48234.map.gz emd_48234.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48234-v30.xml emd-48234-v30.xml emd-48234.xml emd-48234.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48234_fsc.xml emd_48234_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_48234.png emd_48234.png | 140 KB | ||
| Masks |  emd_48234_msk_1.map emd_48234_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-48234.cif.gz emd-48234.cif.gz | 7 KB | ||
| Others |  emd_48234_half_map_1.map.gz emd_48234_half_map_1.map.gz emd_48234_half_map_2.map.gz emd_48234_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48234 http://ftp.pdbj.org/pub/emdb/structures/EMD-48234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48234 | HTTPS FTP |
-Related structure data
| Related structure data |  9mflMC  9mffC  9mfmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_48234.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48234.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map for structure of zebrafish OTOP1 in nanodisc in the presence of inhibitor C11 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_48234_msk_1.map emd_48234_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B for structure of zebrafish OTOP1...
| File | emd_48234_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for structure of zebrafish OTOP1 in nanodisc in the presence of inhibitor C11 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A for structure of zebrafish OTOP1...
| File | emd_48234_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for structure of zebrafish OTOP1 in nanodisc in the presence of inhibitor C11 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : zebrafish OTOP1 in MSP2N2 nanodisc
| Entire | Name: zebrafish OTOP1 in MSP2N2 nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: zebrafish OTOP1 in MSP2N2 nanodisc
| Supramolecule | Name: zebrafish OTOP1 in MSP2N2 nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 131 KDa |
-Macromolecule #1: Proton channel OTOP1
| Macromolecule | Name: Proton channel OTOP1 / type: protein_or_peptide / ID: 1 Details: first two residues (GP) are leftover residues from a 3C protease cleavage site. 3rd residue (V) corresponds to the 2nd residue of the zebrafish OTOP1 sequence (Uniprot:Q7ZWK8) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.817 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPVEHGGTDS MWLNKYNPAA ASSASSSSSS DAENKLFSRL KVSLTKKYPQ KNAELLSAQY GTNLLLLGVS VMLALAAQSG PVKEEHLLS FITVLMLVQL VWMLCYMIRR ERERSPVPER DAHAGASWIR GGLTMLALLS LIMDAFRIGY FVGYHSCISA A LGVYPIVH ...String: GPVEHGGTDS MWLNKYNPAA ASSASSSSSS DAENKLFSRL KVSLTKKYPQ KNAELLSAQY GTNLLLLGVS VMLALAAQSG PVKEEHLLS FITVLMLVQL VWMLCYMIRR ERERSPVPER DAHAGASWIR GGLTMLALLS LIMDAFRIGY FVGYHSCISA A LGVYPIVH ALHTISQVHF LWFHIKDVIK KYETFERFGV IHAVFTNLLL WCNGVMSETE HFMHNHRRRL IEMGYANLST VD VQPHCNC TTSVCSMFST SLYYLYPFNI EYHIFVSAML FVMWKNIGRT LDRHSNRKRR STGSTGLLLG PLGGLVALAS SVS VLVVYL IHLEKTEEMH EAAVSMFYYY GVAMMACMCV GSGTGLLVYR MENRPMDTGS NPARTLDTEL LLASSLGSWL MSWC SVVAS VAEAGQKSPS FSWTSLTYSL LLVLEKCIQN LFIVESLYRR HSEEEEDAAA PQVFSVAVPP YDGILNHGYE AHDKH REAE PAAGSHALSR KQPDAPLPAG QRLDVTPGRK RQILKNICMF LFMCNISLWI LPAFGCRPQY DNPLENETFG TSVWTT VLN VAIPLNLFYR MHSVASLFEV FRKV UniProtKB: Proton channel OTOP1 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: (3beta,5beta,14beta,17alpha)-cholestan-3-ol
| Macromolecule | Name: (3beta,5beta,14beta,17alpha)-cholestan-3-ol / type: ligand / ID: 3 / Number of copies: 4 / Formula: QNJ |
|---|---|
| Molecular weight | Theoretical: 388.669 Da |
| Chemical component information | 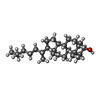 ChemComp-QNJ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||
| Details | sample was complexed with small molecule C11, reaching a final concentration of 1.11mM, and 1% DMSO |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Number real images: 3552 / Average exposure time: 9.8 sec. / Average electron dose: 50.37 e/Å2 Details: Forty-nine frames of 200 ms exposure time were collected per movie |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































