[English] 日本語
 Yorodumi
Yorodumi- EMDB-48111: Human M5 muscarinic acetylcholine receptor complex with mini-Gq a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human M5 muscarinic acetylcholine receptor complex with mini-Gq and iperoxo | |||||||||
 Map data Map data | Human M5 muscarinic acetylcholine receptor complex with mini-Gq and iperoxo | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein-coupled receptor / acetylcholine binding / seven transmembrane protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of phosphatidylinositol dephosphorylation / gastric acid secretion / Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / dopamine transport / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors ...regulation of phosphatidylinositol dephosphorylation / gastric acid secretion / Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / phosphatidylinositol-4,5-bisphosphate phospholipase C activity / dopamine transport / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / photoreceptor outer segment membrane / spectrin binding / alkylglycerophosphoethanolamine phosphodiesterase activity / transmission of nerve impulse / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / cardiac muscle cell apoptotic process / photoreceptor inner segment / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / cell body / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / cellular response to hypoxia / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / postsynaptic membrane / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / GTPase activity / dendrite / synapse / protein-containing complex binding / extracellular exosome / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Burger WAC / Mobbs JI / Thal DM | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Cryo-EM reveals an extrahelical allosteric binding site at the M mAChR. Authors: Wessel A C Burger / Jesse I Mobbs / Bhavika Rana / Jinan Wang / Keya Joshi / Patrick R Gentry / Mahmuda Yeasmin / Hariprasad Venugopal / Aaron M Bender / Craig W Lindsley / Yinglong Miao / ...Authors: Wessel A C Burger / Jesse I Mobbs / Bhavika Rana / Jinan Wang / Keya Joshi / Patrick R Gentry / Mahmuda Yeasmin / Hariprasad Venugopal / Aaron M Bender / Craig W Lindsley / Yinglong Miao / Arthur Christopoulos / Celine Valant / David M Thal /   Abstract: The M muscarinic acetylcholine receptor (M mAChR) represents a promising therapeutic target for neurological disorders. However, the high conservation of its orthosteric binding site poses ...The M muscarinic acetylcholine receptor (M mAChR) represents a promising therapeutic target for neurological disorders. However, the high conservation of its orthosteric binding site poses significant challenges for drug development. While selective positive allosteric modulators (PAMs) offer a potential solution, a structural understanding of the M mAChR and its allosteric binding sites remains limited. Here, we present a 2.8 Å cryo-electron microscopy structure of the M mAChR complexed with heterotrimeric G protein and the agonist iperoxo, completing the active-state structural characterization of the mAChR family. To identify the binding site of M-selective PAMs, we implement an integrated approach combining mutagenesis, pharmacological assays, structural biology, and molecular dynamics simulations. Our mutagenesis studies reveal that selective M PAMs bind outside previously characterized M mAChR allosteric sites. Subsequently, we obtain a 2.1 Å structure of M mAChR co-bound with acetylcholine and the selective PAM VU6007678, revealing an allosteric pocket at the extrahelical interface between transmembrane domains 3 and 4 that is confirmed through mutagenesis and simulations. These findings demonstrate the diverse mechanisms of allosteric regulation in mAChRs and highlight the value of integrating pharmacological and structural approaches to identify allosteric binding sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48111.map.gz emd_48111.map.gz | 51 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48111-v30.xml emd-48111-v30.xml emd-48111.xml emd-48111.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48111_fsc.xml emd_48111_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_48111.png emd_48111.png | 99.7 KB | ||
| Masks |  emd_48111_msk_1.map emd_48111_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-48111.cif.gz emd-48111.cif.gz | 6.7 KB | ||
| Others |  emd_48111_half_map_1.map.gz emd_48111_half_map_1.map.gz emd_48111_half_map_2.map.gz emd_48111_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48111 http://ftp.pdbj.org/pub/emdb/structures/EMD-48111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48111 | HTTPS FTP |
-Related structure data
| Related structure data |  9ek0MC  9ejzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_48111.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48111.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human M5 muscarinic acetylcholine receptor complex with mini-Gq and iperoxo | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_48111_msk_1.map emd_48111_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_48111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_48111_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human M5-miniGq-Gb1g2 complex
| Entire | Name: Human M5-miniGq-Gb1g2 complex |
|---|---|
| Components |
|
-Supramolecule #1: Human M5-miniGq-Gb1g2 complex
| Supramolecule | Name: Human M5-miniGq-Gb1g2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Muscarinic acetylcholine receptor M5
| Macromolecule | Name: Muscarinic acetylcholine receptor M5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.190051 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAEG DSYHNATTVN GTPVNHQPLE RHRLWEVITI AAVTAVVSLI TIVGNVLVMI SFKVNSQLKT VNNYYLLSLA CADLIIGIF SMNLYTTYIL MGRWALGSLA CDLWLALDYV ASNASVMNLL VISFDRYFSI TRPLTYRAKR TPKRAGIMIG L AWLISFIL ...String: DYKDDDDAEG DSYHNATTVN GTPVNHQPLE RHRLWEVITI AAVTAVVSLI TIVGNVLVMI SFKVNSQLKT VNNYYLLSLA CADLIIGIF SMNLYTTYIL MGRWALGSLA CDLWLALDYV ASNASVMNLL VISFDRYFSI TRPLTYRAKR TPKRAGIMIG L AWLISFIL WAPAILCWQY LVGKRTVPLD ECQIQFLSEP TITFGTAIAA FYIPVSVMTI LYCRIYRETE KRTKDLADLQ GS DSVPSHQ MTKRKRVVLV KERKAAQTLS AILLAFIITW TPYNIMVLVS TFCDKCVPVT LWHLGYWLCY VNSTVNPICY ALC NRTFRK TFKMLLLCRW KKKKVEE UniProtKB: Muscarinic acetylcholine receptor M5, Muscarinic acetylcholine receptor M5 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein Gq alpha subunit chimera
| Macromolecule | Name: Guanine nucleotide-binding protein Gq alpha subunit chimera type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.039793 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GCTLSAEDKA AVERSKMIDR NLREDGEKAR RTLRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR ...String: GCTLSAEDKA AVERSKMIDR NLREDGEKAR RTLRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR YTTPEDATPE PGEDPRVTRA KYFIRKEFVD ISTASGDGRH ICYPHFTCAV DTENARRIFN DCKDIILQMN LR EYNLV |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium
| Macromolecule | Name: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium type: ligand / ID: 5 / Number of copies: 1 / Formula: IXO |
|---|---|
| Molecular weight | Theoretical: 197.254 Da |
| Chemical component information | 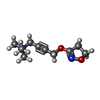 ChemComp-IXO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































