[English] 日本語
 Yorodumi
Yorodumi- EMDB-48048: Structure of the prefusion HKU5-19s Spike trimer (conformation 2) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the prefusion HKU5-19s Spike trimer (conformation 2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MERS-related HKU5 coronaviruses / MERSr-CoV / Spike glycoprotein / fusion protein / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Pipistrellus bat coronavirus HKU5 Pipistrellus bat coronavirus HKU5 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Park YJ / Gen R / Seattle Structural Genomics Center for Infectious Disease (SSGCID) / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2025 Journal: Cell / Year: 2025Title: Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses. Authors: Young-Jun Park / Chen Liu / Jimin Lee / Jack T Brown / Cheng-Bao Ma / Peng Liu / Risako Gen / Qing Xiong / Samantha K Zepeda / Cameron Stewart / Amin Addetia / Caroline J Craig / M Alejandra ...Authors: Young-Jun Park / Chen Liu / Jimin Lee / Jack T Brown / Cheng-Bao Ma / Peng Liu / Risako Gen / Qing Xiong / Samantha K Zepeda / Cameron Stewart / Amin Addetia / Caroline J Craig / M Alejandra Tortorici / Abeer N Alshukairi / Tyler N Starr / Huan Yan / David Veesler /    Abstract: DPP4 was considered a canonical receptor for merbecoviruses until the recent discovery of African bat-borne MERS-related coronaviruses using ACE2. The extent and diversity of ACE2 utilization among ...DPP4 was considered a canonical receptor for merbecoviruses until the recent discovery of African bat-borne MERS-related coronaviruses using ACE2. The extent and diversity of ACE2 utilization among merbecoviruses and their receptor species tropism remain unknown. Here, we reveal that HKU5 enters host cells utilizing Pipistrellus abramus (P.abr) and several non-bat mammalian ACE2s through a binding mode distinct from that of any other known ACE2-using coronaviruses. We defined the molecular determinants of receptor species tropism and identified a single amino acid mutation enabling HKU5 to utilize human ACE2, providing proof of principle for machine-learning-assisted outbreak preparedness. We show that MERS-CoV and HKU5 have markedly distinct antigenicity and identified several HKU5 inhibitors, including two clinical compounds. Our findings profoundly alter our understanding of coronavirus evolution, as several merbecovirus clades independently evolved ACE2 utilization, and pave the way for developing countermeasures against viruses poised for human emergence. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48048.map.gz emd_48048.map.gz | 810.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48048-v30.xml emd-48048-v30.xml emd-48048.xml emd-48048.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_48048.png emd_48048.png | 104.8 KB | ||
| Filedesc metadata |  emd-48048.cif.gz emd-48048.cif.gz | 7.2 KB | ||
| Others |  emd_48048_additional_1.map.gz emd_48048_additional_1.map.gz emd_48048_half_map_1.map.gz emd_48048_half_map_1.map.gz emd_48048_half_map_2.map.gz emd_48048_half_map_2.map.gz | 431.1 MB 795.2 MB 795.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48048 http://ftp.pdbj.org/pub/emdb/structures/EMD-48048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48048 | HTTPS FTP |
-Related structure data
| Related structure data |  9eh8MC  9d32C  9e0iC  9ea0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_48048.map.gz / Format: CCP4 / Size: 857.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48048.map.gz / Format: CCP4 / Size: 857.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.843 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_48048_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_48048_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_48048_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion HKU5-19s S trimer
| Entire | Name: Prefusion HKU5-19s S trimer |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion HKU5-19s S trimer
| Supramolecule | Name: Prefusion HKU5-19s S trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pipistrellus bat coronavirus HKU5 Pipistrellus bat coronavirus HKU5 |
| Molecular weight | Theoretical: 150.750672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQSVDMGTT GSGNCIESQV QPDFFETARN TWPLSIDTSK AEGVIYPNGK SYSNITLTY TGLYPKANDL GKQYVFSDGH SSPGTLSRLF VSNYSRQVEP FDSGFVVRIG AAANKTGTTI ISQSTNRPIK K IYPAFMLG ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQSVDMGTT GSGNCIESQV QPDFFETARN TWPLSIDTSK AEGVIYPNGK SYSNITLTY TGLYPKANDL GKQYVFSDGH SSPGTLSRLF VSNYSRQVEP FDSGFVVRIG AAANKTGTTI ISQSTNRPIK K IYPAFMLG HSVGNYTPTN ITGRYLNHTL VILPDGCGTL VHAFYCILQP RTQAYCAGAS TFTSVTVWDT PASDCANSQS YN RLANLNA FKLYFDLINC TFRYNYTITE DENAEWFGIT QDTQGVHLYS SRKENVFRNN MFHFATLPVY QKILYYTVIP RSI RSPFND RKAWAAFYIY KLHPLTYLLN FDVEGYITKA VDCGYDDLAQ LQCSYQSFDV ETGVYSVSSF EASPRGEFIE QATT QECDF TPMLTGTPPP IYNFKRLVFT NCNYNLTKLL SLFQVSEFSC HQVSPSSLAT GCYSSLTVDY FAYSTDMSSY LQPGS AGEI VQFNYKQDFS NPTCRVLATV PQNLTTITKP SNYAYLTECY KTSAYGKNYL YNAPGGYTPC LSLASRGFST KYQSHS DGE LTTTGYIYPV TGNLQMAFII SVQYGTDTNS VCPMQALRND TSIEDKLDVC VEYSLHGITG RGVFHNCTSV GLRNQRF VY DTFDNLVGYH SYNGNYYCVR PCVSVPVSVI YDKVSNSHAT LFGSVACSHV TTMMSQFSRM TKTNLLARTT PGPLQTVV G CAMGFINTSM VVDECQLPLG QSLCAIPPNP SARLARASSG VTDVFQIATL NFTSPLTLAP INSTGFVVAV PTNFTFGVT QEYIETTIQK ITVDCKQYVC NGFKKCEELL TEYGQFCSKI NQALHGANLR QDESIANLFS SIKTQNTQPL QAGLNGDFNL TMLQIPQVT TGERKYRSAI EDLLFNKVTI ADPGYMQGYD ECMQQGPQSA RDLICAQYVA GYKVLPPLYD PYMEAAYTSS L LGSIAGAS WTAGLSSFAA IPFAQSIFYR LNGVGITQQV LSENQKIIAN KFNQALGAMQ TGFTTTNLAF NKVQDAVNAN AM ALSKLAA ELSNTFGAIS SSISDILARL DPPEQEAQID RLINGRLTSL NAFVAQQLVR TEAAARSAQL AQDKVNECVK SQS KRNGFC GTGTHIVSFA INAPNGLYFF HVGYQPTSHV NATAAYGLCN TENPPKCIAP IGGYFVLNQT TSTVANSEQQ WYYT GSSFF HPEPITEVNS KYVSMDVKFE NLTNKLPPPL LSNTTDLDFK EELEEFFKNV SSQGPNFQEI SKINTTLLNL NTELM VLSE VVKQLNESYI DLKELGNYTF YQKGSGYIPE APRDGQAYVR KDGEWVLLST FLGGSGLNDI FEAQKIEWHE GGSHHH HHH HH UniProtKB: Spike glycoprotein |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: FOLIC ACID
| Macromolecule | Name: FOLIC ACID / type: ligand / ID: 8 / Number of copies: 3 / Formula: FOL |
|---|---|
| Molecular weight | Theoretical: 441.397 Da |
| Chemical component information | 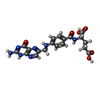 ChemComp-FA: |
-Macromolecule #9: LINOLEIC ACID
| Macromolecule | Name: LINOLEIC ACID / type: ligand / ID: 9 / Number of copies: 3 / Formula: EIC |
|---|---|
| Molecular weight | Theoretical: 280.445 Da |
| Chemical component information |  ChemComp-EIC: |
-Macromolecule #10: water
| Macromolecule | Name: water / type: ligand / ID: 10 / Number of copies: 813 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































