[English] 日本語
 Yorodumi
Yorodumi- EMDB-47132: Human GABAA receptor of beta2-alpha1-beta2-alpha1-gamma2 subtype ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human GABAA receptor of beta2-alpha1-beta2-alpha1-gamma2 subtype in complex with GABA plus Lamotrigine | |||||||||
 Map data Map data | The map with better TMD density. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channels / Heteropentamer / Receptor / Inhibitory. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbenzodiazepine receptor activity / GABA receptor complex / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation ...benzodiazepine receptor activity / GABA receptor complex / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / chloride channel activity / adult behavior / Signaling by ERBB4 / cochlea development / chloride channel complex / dendrite membrane / cytoplasmic vesicle membrane / chloride transmembrane transport / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / chemical synaptic transmission / dendritic spine / postsynaptic membrane / postsynapse / axon / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Zhou J / Hibbs RE / Noviello CM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Resolving native GABA receptor structures from the human brain. Authors: Jia Zhou / Colleen M Noviello / Jinfeng Teng / Haley Moore / Bradley Lega / Ryan E Hibbs /  Abstract: Type A GABA (γ-aminobutyric acid) receptors (GABA receptors) mediate most fast inhibitory signalling in the brain and are targets for drugs that treat epilepsy, anxiety, depression and insomnia and ...Type A GABA (γ-aminobutyric acid) receptors (GABA receptors) mediate most fast inhibitory signalling in the brain and are targets for drugs that treat epilepsy, anxiety, depression and insomnia and for anaesthetics. These receptors comprise a complex array of 19 related subunits, which form pentameric ligand-gated ion channels. The composition and structure of native GABA receptors in the human brain have been inferred from subunit localization in tissue, functional measurements and structural analysis from recombinant expression and in mice. However, the arrangements of subunits that co-assemble physiologically in native human GABA receptors remain unknown. Here we isolated α1 subunit-containing GABA receptors from human patients with epilepsy. Using cryo-electron microscopy, we defined a set of 12 native subunit assemblies and their 3D structures. We address inconsistencies between previous native and recombinant approaches, and reveal details of previously undefined subunit interfaces. Drug-like densities in a subset of these interfaces led us to uncover unexpected activity on the GABA receptor of antiepileptic drugs and resulted in localization of one of these drugs to the benzodiazepine-binding site. Proteomics and further structural analysis suggest interactions with the auxiliary subunits neuroligin 2 and GARLH4, which localize and modulate GABA receptors at inhibitory synapses. This work provides a structural foundation for understanding GABA receptor signalling and targeted pharmacology in the human brain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47132.map.gz emd_47132.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47132-v30.xml emd-47132-v30.xml emd-47132.xml emd-47132.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_47132.png emd_47132.png | 120.6 KB | ||
| Masks |  emd_47132_msk_1.map emd_47132_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-47132.cif.gz emd-47132.cif.gz | 7.8 KB | ||
| Others |  emd_47132_additional_1.map.gz emd_47132_additional_1.map.gz emd_47132_half_map_1.map.gz emd_47132_half_map_1.map.gz emd_47132_half_map_2.map.gz emd_47132_half_map_2.map.gz | 59.6 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47132 http://ftp.pdbj.org/pub/emdb/structures/EMD-47132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47132 | HTTPS FTP |
-Validation report
| Summary document |  emd_47132_validation.pdf.gz emd_47132_validation.pdf.gz | 939.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_47132_full_validation.pdf.gz emd_47132_full_validation.pdf.gz | 938.8 KB | Display | |
| Data in XML |  emd_47132_validation.xml.gz emd_47132_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_47132_validation.cif.gz emd_47132_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47132 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47132 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47132 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47132 | HTTPS FTP |
-Related structure data
| Related structure data |  9drxMC  9crsC  9crvC  9csbC  9ct0C  9ctjC  9ctpC  9ctvC  9cx7C  9cxaC  9cxbC  9cxcC  9cxdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47132.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47132.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map with better TMD density. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_47132_msk_1.map emd_47132_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: The map with better ECD density.
| File | emd_47132_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map with better ECD density. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_47132_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_47132_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Recombinant GABAA receptor complex with Lamotrigine
| Entire | Name: Recombinant GABAA receptor complex with Lamotrigine |
|---|---|
| Components |
|
-Supramolecule #1: Recombinant GABAA receptor complex with Lamotrigine
| Supramolecule | Name: Recombinant GABAA receptor complex with Lamotrigine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gamma-aminobutyric acid receptor subunit beta-2
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit beta-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.810086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVNDPSNMS LVKETVDRLL KGYDIRLRPD FGGPPVAVGM NIDIASIDMV SEVNMDYTLT MYFQQAWRDK RLSYNVIPLN LTLDNRVAD QLWVPDTYFL NDKKSFVHGV TVKNRMIRLH PDGTVLYGLR ITTTAACMMD LRRYPLDEQN CTLEIESYGY T TDDIEFYW ...String: QSVNDPSNMS LVKETVDRLL KGYDIRLRPD FGGPPVAVGM NIDIASIDMV SEVNMDYTLT MYFQQAWRDK RLSYNVIPLN LTLDNRVAD QLWVPDTYFL NDKKSFVHGV TVKNRMIRLH PDGTVLYGLR ITTTAACMMD LRRYPLDEQN CTLEIESYGY T TDDIEFYW RGDDNAVTGV TKIELPQFSI VDYKLITKKV VFSTGSYPRL SLSFKLKRNI GYFILQTYMP SILITILSWV SF WINYDAS AARVALGITT VLTMTTINTH LRETLPKIPY VKAIDMYLMG CFVFVFMALL EYALVNYIFF SQPARAAAID RWS RIFFPV VFSFFNIVYW LYYVNVDGSG ATNFSLLKQA GDVEENPG UniProtKB: Gamma-aminobutyric acid receptor subunit beta-2, Gamma-aminobutyric acid receptor subunit beta-2 |
-Macromolecule #2: Gamma-aminobutyric acid receptor subunit alpha-1
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit alpha-1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.061211 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QPSLQDELKD NTTVFTRILD RLLDGYDNRL RPGLGERVTE VKTDIFVTSF GPVSDHDMEY TIDVFFRQSW KDERLKFKGP MTVLRLNNL MASKIWTPDT FFHNGKKSVA HNMTMPNKLL RITEDGTLLY TMRLTVRAEC PMHLEDFPMD AHACPLKFGS Y AYTRAEVV ...String: QPSLQDELKD NTTVFTRILD RLLDGYDNRL RPGLGERVTE VKTDIFVTSF GPVSDHDMEY TIDVFFRQSW KDERLKFKGP MTVLRLNNL MASKIWTPDT FFHNGKKSVA HNMTMPNKLL RITEDGTLLY TMRLTVRAEC PMHLEDFPMD AHACPLKFGS Y AYTRAEVV YEWTREPARS VVVAEDGSRL NQYDLLGQTV DSGIVQSSTG EYVVMTTHFH LKRKIGYFVI QTYLPCIMTV IL SQVSFWL NRESVPARTV FGVTTVLTMT TLSISARNSL PKVAYATAMD WFIAVCYAFV FSALIEFATV NYFTKSQPAR AAK IDRLSR IAFPLLFGIF NLVYWATYLN REPQLKAPTP HQ UniProtKB: Gamma-aminobutyric acid receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit alpha-1 |
-Macromolecule #3: Gamma-aminobutyric acid receptor subunit gamma-2
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit gamma-2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.673109 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: WSHPQFEKGG GSGGGSGGSS AWSHPQFEKL EVLFQGPQKS DDDYEDYASN KTWVLTPKVP EGDVTVILNN LLEGYDNKLR PDIGVKPTL IHTDMYVNSI GPVNAINMEY TIDIFFAQTW YDRRLKFNST IKVLRLNSNM VGKIWIPDTF FRNSKKADAH W ITTPNRML ...String: WSHPQFEKGG GSGGGSGGSS AWSHPQFEKL EVLFQGPQKS DDDYEDYASN KTWVLTPKVP EGDVTVILNN LLEGYDNKLR PDIGVKPTL IHTDMYVNSI GPVNAINMEY TIDIFFAQTW YDRRLKFNST IKVLRLNSNM VGKIWIPDTF FRNSKKADAH W ITTPNRML RIWNDGRVLY TLRLTIDAEC QLQLHNFPMD EHSCPLEFSS YGYPREEIVY QWKRSSVEVG DTRSWRLYQF SF VGLRNTT EVVKTTSGDY VVMSVYFDLS RRMGYFTIQT YIPCTLIVVL SWVSFWINKD AVPARTSLGI TTVLTMTTLS TIA RKSLPK VSYVTAMDLF VSVCFIFVFS ALVEYGTLHY FVSSQPARAA KMDSYARIFF PTAFCLFNLV YWVSYLYLSR GSGA TNFSL LKQAGDVEEN PG UniProtKB: Gamma-aminobutyric acid receptor subunit gamma-2, Gamma-aminobutyric acid receptor subunit gamma-2 |
-Macromolecule #4: Kappa Fab 1F4 Light Chain
| Macromolecule | Name: Kappa Fab 1F4 Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.505943 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NIVMTQSPKS MSMSVGERVT LSCKASEYVG TYVSWYQQKP EQSPKLLIYG ASNRYTGVPD RFTGSGSATD FTLTIGSVQA EDLADYHCG QSYSYPTFGA GTKLELKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK ...String: NIVMTQSPKS MSMSVGERVT LSCKASEYVG TYVSWYQQKP EQSPKLLIYG ASNRYTGVPD RFTGSGSATD FTLTIGSVQA EDLADYHCG QSYSYPTFGA GTKLELKRAD AAPTVSIFPP SSEQLTSGGA SVVCFLNNFY PKDINVKWKI DGSERQNGVL N SWTDQDSK DSTYSMSSTL TLTKDEYERH NSYTCEATHK TSTSPIVKSF NRNEC |
-Macromolecule #5: IgG2b Fab 1F4 Heavy Chain
| Macromolecule | Name: IgG2b Fab 1F4 Heavy Chain / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.811043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQQSGAE LVKPGASVKL SCTASGFNIK DTYMYWVKQR PEQGLEWIGR IDPANGDTKY DPKFQGKATI TTDTFSNTAY LQLSSLTSE DTAVYYCARK GLRWAMDYWG QGTSVTVSTA KTTPPSVYPL APGCGDTTGS SVTLGCLVKG YFPESVTVTW N SGSLSSSV ...String: EVQLQQSGAE LVKPGASVKL SCTASGFNIK DTYMYWVKQR PEQGLEWIGR IDPANGDTKY DPKFQGKATI TTDTFSNTAY LQLSSLTSE DTAVYYCARK GLRWAMDYWG QGTSVTVSTA KTTPPSVYPL APGCGDTTGS SVTLGCLVKG YFPESVTVTW N SGSLSSSV HTFPALLQSG LYTMSSSVTV PSSTWPSQTV TCSVAHPASS TTVDKKLEPS GPISTINPCP PCKECHKCPA PN LEGGPSV FIFPPNIKDV LMISLTPKVT CVVVDVSEDD PDVQISWFVN NVEVHTAQTQ THREDYNSTI RVVSTLPIQH QDW MSGKEF KCKVNNKDLP SPIERTISKI KGLVRAPQVY ILPPPAEQLS RKDVSLTCLV VGFNPGDISV EWTSNGHTEE NYKD TAPVL DSDGSYFIYS KLNMKTSKWE KTDSFSCNVR HEGLKNYYLK KTISRSPGK |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #11: GAMMA-AMINO-BUTANOIC ACID
| Macromolecule | Name: GAMMA-AMINO-BUTANOIC ACID / type: ligand / ID: 11 / Number of copies: 2 / Formula: ABU |
|---|---|
| Molecular weight | Theoretical: 103.12 Da |
| Chemical component information |  ChemComp-ABU: |
-Macromolecule #12: (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine
| Macromolecule | Name: (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine type: ligand / ID: 12 / Number of copies: 1 / Formula: IYJ |
|---|---|
| Molecular weight | Theoretical: 256.091 Da |
| Chemical component information | 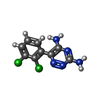 ChemComp-IYJ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV / Details: 3.5 s Blot.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































