+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial ClpP in complex with Bortezomib | |||||||||
 Map data Map data | D7 symmetry map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protease / Proteolysis / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane protein proteolysis / mitochondrial protein catabolic process / endopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / Mitochondrial protein degradation / : / peptidase activity / ATPase binding ...membrane protein proteolysis / mitochondrial protein catabolic process / endopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / Mitochondrial protein degradation / : / peptidase activity / ATPase binding / endopeptidase activity / mitochondrial matrix / serine-type endopeptidase activity / mitochondrion / proteolysis / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.49 Å | |||||||||
 Authors Authors | Uday AB / Zeytuni N / Goncalves M / Vahidi S | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Mechanism of allosteric activation in human mitochondrial ClpP protease. Authors: Monica M Goncalves / Adwaith B Uday / Taylor J B Forrester / S Quinn W Currie / Angelina S Kim / Yue Feng / Yulia Jitkova / Algirdas Velyvis / Robert W Harkness / Matthew S Kimber / Aaron D ...Authors: Monica M Goncalves / Adwaith B Uday / Taylor J B Forrester / S Quinn W Currie / Angelina S Kim / Yue Feng / Yulia Jitkova / Algirdas Velyvis / Robert W Harkness / Matthew S Kimber / Aaron D Schimmer / Natalie Zeytuni / Siavash Vahidi /  Abstract: Human ClpP protease contributes to mitochondrial protein quality control by degrading misfolded proteins. ClpP is overexpressed in cancers such as acute myeloid leukemia (AML), where its inhibition ...Human ClpP protease contributes to mitochondrial protein quality control by degrading misfolded proteins. ClpP is overexpressed in cancers such as acute myeloid leukemia (AML), where its inhibition leads to the accumulation of damaged respiratory chain subunits and cell death. Conversely, hyperactivating ClpP with small-molecule activators, such as the recently discovered ONC201, disrupts mitochondrial protein degradation and impairs respiration in cancer cells. Despite its critical role in human health, the mechanism underlying the structural and functional properties of human ClpP remains elusive. Notably, human ClpP is paradoxically activated by active-site inhibitors. All available structures of human ClpP published to date are in the inactive compact or compressed states, surprisingly even when ClpP is bound to an activator molecule such as ONC201. Here, we present structures of human mitochondrial ClpP in the active extended state, including a pair of structures where ClpP is bound to an active-site inhibitor. We demonstrate that amino acid substitutions in the handle region (A192E and E196R) recreate a conserved salt bridge found in bacterial ClpP, stabilizing the extended active state and significantly enhancing ClpP activity. We elucidate the ClpP activation mechanism, highlighting a hormetic effect where substoichiometric inhibitor binding triggers an allosteric transition that drives ClpP into its active extended state. Our findings link the conformational dynamics of ClpP to its catalytic function and provide high-resolution structures for the rational design of potent and specific ClpP inhibitors, with implications for targeting AML and other disorders with ClpP involvement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_46971.map.gz emd_46971.map.gz | 106.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-46971-v30.xml emd-46971-v30.xml emd-46971.xml emd-46971.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_46971_fsc.xml emd_46971_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_46971.png emd_46971.png | 75.1 KB | ||
| Filedesc metadata |  emd-46971.cif.gz emd-46971.cif.gz | 6.1 KB | ||
| Others |  emd_46971_half_map_1.map.gz emd_46971_half_map_1.map.gz emd_46971_half_map_2.map.gz emd_46971_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-46971 http://ftp.pdbj.org/pub/emdb/structures/EMD-46971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46971 | HTTPS FTP |
-Related structure data
| Related structure data |  9dkwMC  9dkvC  9dqkC  9dqlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_46971.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_46971.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D7 symmetry map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: D7 symmetry half map
| File | emd_46971_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D7 symmetry half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: D7 symmetry half map
| File | emd_46971_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | D7 symmetry half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial ClpP in complex with Bortezomib
| Entire | Name: Human mitochondrial ClpP in complex with Bortezomib |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial ClpP in complex with Bortezomib
| Supramolecule | Name: Human mitochondrial ClpP in complex with Bortezomib / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-dependent Clp protease proteolytic subunit, mitochondrial
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.179875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SLIPIVVEQT GRGERAYDIY SRLLRERIVC VMGPIDDSVA SLVIAQLLFL QSESNKKPIH MYINSPGGVV TAGLAIYDTM QYILNPICT WCVGQAASMG SLLLAAGTPG MRHSLPNSRI MIHQPSGGAR GQATDIAIQA EEIMKLKKQL YNIYAKHTKQ S LQVIESAM ...String: SLIPIVVEQT GRGERAYDIY SRLLRERIVC VMGPIDDSVA SLVIAQLLFL QSESNKKPIH MYINSPGGVV TAGLAIYDTM QYILNPICT WCVGQAASMG SLLLAAGTPG MRHSLPNSRI MIHQPSGGAR GQATDIAIQA EEIMKLKKQL YNIYAKHTKQ S LQVIESAM ERDRYMSPME AQEFGILDKV LVHPPQDGED EPTLVQKEPV EAAPAAEPVP AST UniProtKB: ATP-dependent Clp protease proteolytic subunit, mitochondrial |
-Macromolecule #2: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL...
| Macromolecule | Name: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE type: ligand / ID: 2 / Number of copies: 14 / Formula: BO2 |
|---|---|
| Molecular weight | Theoretical: 384.237 Da |
| Chemical component information | 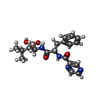 ChemComp-BO2: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 168 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 18 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































