+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4642 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 12 Angstrom structure of detergent solubilised LAT1-CD98hc | |||||||||
 Map data Map data | 12 angstrom structure of LAT1-CD98hc | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Chiduza GN / Johnson R / Wright GS / Antonyuk S / Muench S / Hasnain SS | |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2019 Journal: Acta Crystallogr D Struct Biol / Year: 2019Title: LAT1 (SLC7A5) and CD98hc (SLC3A2) complex dynamics revealed by single-particle cryo-EM. Authors: George N Chiduza / Rachel M Johnson / Gareth S A Wright / Svetlana V Antonyuk / Stephen P Muench / S Samar Hasnain /  Abstract: Solute carriers are a large class of transporters that play key roles in normal and disease physiology. Among the solute carriers, heteromeric amino-acid transporters (HATs) are unique in their ...Solute carriers are a large class of transporters that play key roles in normal and disease physiology. Among the solute carriers, heteromeric amino-acid transporters (HATs) are unique in their quaternary structure. LAT1-CD98hc, a HAT, transports essential amino acids and drugs across the blood-brain barrier and into cancer cells. It is therefore an important target both biologically and therapeutically. During the course of this work, cryo-EM structures of LAT1-CD98hc in the inward-facing conformation and in either the substrate-bound or apo states were reported to 3.3-3.5 Å resolution [Yan et al. (2019), Nature (London), 568, 127-130]. Here, these structures are analyzed together with our lower resolution cryo-EM structure, and multibody 3D auto-refinement against single-particle cryo-EM data was used to characterize the dynamics of the interaction of CD98hc and LAT1. It is shown that the CD98hc ectodomain and the LAT1 extracellular surface share no substantial interface. This allows the CD98hc ectodomain to have a high degree of movement within the extracellular space. The functional implications of these aspects are discussed together with the structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4642.map.gz emd_4642.map.gz | 3.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4642-v30.xml emd-4642-v30.xml emd-4642.xml emd-4642.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
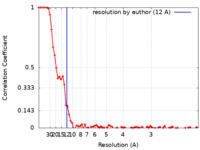
| FSC (resolution estimation) |  emd_4642_fsc.xml emd_4642_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4642.png emd_4642.png | 75.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4642 http://ftp.pdbj.org/pub/emdb/structures/EMD-4642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4642 | HTTPS FTP |
-Validation report
| Summary document |  emd_4642_validation.pdf.gz emd_4642_validation.pdf.gz | 222.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4642_full_validation.pdf.gz emd_4642_full_validation.pdf.gz | 221.9 KB | Display | |
| Data in XML |  emd_4642_validation.xml.gz emd_4642_validation.xml.gz | 9.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4642 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4642 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4642 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4642 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4642.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4642.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 12 angstrom structure of LAT1-CD98hc | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : LAT1-CD98hc
| Entire | Name: LAT1-CD98hc |
|---|---|
| Components |
|
-Supramolecule #1: LAT1-CD98hc
| Supramolecule | Name: LAT1-CD98hc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Location in cell: Plasma membrane Homo sapiens (human) / Location in cell: Plasma membrane |
| Molecular weight | Theoretical: 123 kDa/nm |
-Macromolecule #1: LAT1
| Macromolecule | Name: LAT1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGAGPKRRA LAAPAAEEKE EAREKMLAAK SADGSAPAGE GEGVTLQRNI TLLNGVAIIV GTIIGSGIF VTPTGVLKEA GSPGLALVVW AACGVFSIVG ALCYAELGTT ISKSGGDYAY M LEVYGSLP AFLKLWIELL IIRPSSQYIV ALVFATYLLK PLFPTCPVPE ...String: MAGAGPKRRA LAAPAAEEKE EAREKMLAAK SADGSAPAGE GEGVTLQRNI TLLNGVAIIV GTIIGSGIF VTPTGVLKEA GSPGLALVVW AACGVFSIVG ALCYAELGTT ISKSGGDYAY M LEVYGSLP AFLKLWIELL IIRPSSQYIV ALVFATYLLK PLFPTCPVPE EAAKLVACLC VL LLTAVNC YSVKAATRVQ DAFAAAKLLA LALIILLGFV QIGKGDVSNL DPNFSFEGTK LDV GNIVLA LYSGLFAYGG WNYLNFVTEE MINPYRNLPL AIIISLPIVT LVYVLTNLAY FTTL STEQM LSSEAVAVDF GNYHLGVMSW IIPVFVGLSC FGSVNGSLFT SSRLFFVGSR EGHLP SILS MIHPQLLTPV PSLVFTCVMT LLYAFSKDIF SVINFFSFFN WLCVALAIIG MIWLRH RKP ELERPIKVNL ALPVFFILAC LFLIAVSFWK TPVECGIGFT IILSGLPVYF FGVWWKN KP KWLLQGIFST TVLCQKLMQV VPQET |
-Macromolecule #2: CD98hc
| Macromolecule | Name: CD98hc / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELQPPEASI AVVSIPRQLP GSHSEAGVQG LSAGDDSELG SHCVAQTGLE LLASGDPLPS ASQNAEMIE TGSDCVTQAG LQLLASSDPP ALASKNAEVT GTMSQDTEVD MKEVELNELE P EKQPMNAA SGAAMSLAGA EKNGLVKIKV AEDEAEAAAA AKFTGLSKEE ...String: MELQPPEASI AVVSIPRQLP GSHSEAGVQG LSAGDDSELG SHCVAQTGLE LLASGDPLPS ASQNAEMIE TGSDCVTQAG LQLLASSDPP ALASKNAEVT GTMSQDTEVD MKEVELNELE P EKQPMNAA SGAAMSLAGA EKNGLVKIKV AEDEAEAAAA AKFTGLSKEE LLKVAGSPGW VR TRWALLL LFWLGWLGML AGAVVIIVRA PRCRELPAQK WWHTGALYRI GDLQAFQGHG AGN LAGLKG RLDYLSSLKV KGLVLGPIHK NQKDDVAQTD LLQIDPNFGS KEDFDSLLQS AKKK SIRVI LDLTPNYRGE NSWFSTQVDT VATKVKDALE FWLQAGVDGF QVRDIENLKD ASSFL AEWQ NITKGFSEDR LLIAGTNSSD LQQILSLLES NKDLLLTSSY LSDSGSTGEH TKSLVT QYL NATGNRWCSW SLSQARLLTS FLPAQLLRLY QLMLFTLPGT PVFSYGDEIG LDAAALP GQ PMEAPVMLWD ESSFPDIPGA VSANMTVKGQ SEDPGSLLSL FRRLSDQRSK ERSLLHGD F HAFSAGPGLF SYIRHWDQNE RFLVVLNFGD VGLSAGLQAS DLPASASLPA KADLLLSTQ PGREEGSPLE LERLKLEPHE GLLLRFPYAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.3 mg/mL |
|---|---|
| Buffer | pH: 8 Details: Buffer: 100 mM Tris-Cl, 300 mM NaCl, 0.01% w/v DDM/LMNG/CHS (15:3:1), pH8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 4.96 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)