[English] 日本語
 Yorodumi
Yorodumi- EMDB-4611: Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in d... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4611 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lipid scrambles / TMEM16 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium activated galactosylceramide scrambling / phosphatidylserine exposure on blood platelet / calcium activated phosphatidylserine scrambling / calcium activated phosphatidylcholine scrambling / calcium activated phospholipid scrambling / positive regulation of potassium ion export across plasma membrane / positive regulation of monoatomic ion transmembrane transport / activation of blood coagulation via clotting cascade / purinergic nucleotide receptor signaling pathway / phospholipid scramblase activity ...calcium activated galactosylceramide scrambling / phosphatidylserine exposure on blood platelet / calcium activated phosphatidylserine scrambling / calcium activated phosphatidylcholine scrambling / calcium activated phospholipid scrambling / positive regulation of potassium ion export across plasma membrane / positive regulation of monoatomic ion transmembrane transport / activation of blood coagulation via clotting cascade / purinergic nucleotide receptor signaling pathway / phospholipid scramblase activity / negative regulation of cell volume / cholinergic synapse / bone mineralization involved in bone maturation / intracellularly calcium-gated chloride channel activity / pore complex assembly / plasma membrane phospholipid scrambling / voltage-gated monoatomic ion channel activity / positive regulation of phagocytosis, engulfment / bleb assembly / Stimuli-sensing channels / voltage-gated chloride channel activity / calcium-activated cation channel activity / positive regulation of monocyte chemotaxis / chloride transport / dendritic cell chemotaxis / phospholipid translocation / chloride channel activity / regulation of postsynaptic membrane potential / positive regulation of endothelial cell apoptotic process / positive regulation of bone mineralization / chloride channel complex / Neutrophil degranulation / chloride transmembrane transport / synaptic membrane / sodium ion transmembrane transport / calcium ion transmembrane transport / blood coagulation / positive regulation of apoptotic process / protein homodimerization activity / metal ion binding / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
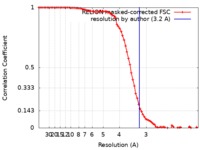
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Alvadia C / Lim NK | |||||||||
| Funding support |  Switzerland, Switzerland,  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Authors: Carolina Alvadia / Novandy K Lim / Vanessa Clerico Mosina / Gert T Oostergetel / Raimund Dutzler / Cristina Paulino /   Abstract: The lipid scramblase TMEM16F initiates blood coagulation by catalyzing the exposure of phosphatidylserine in platelets. The protein is part of a family of membrane proteins, which encompasses calcium- ...The lipid scramblase TMEM16F initiates blood coagulation by catalyzing the exposure of phosphatidylserine in platelets. The protein is part of a family of membrane proteins, which encompasses calcium-activated channels for ions and lipids. Here, we reveal features of murine TMEM16F (mTMEM16F) that underlie its function as a lipid scramblase and an ion channel. The cryo-EM data of mTMEM16F in absence and presence of Ca define the ligand-free closed conformation of the protein and the structure of a Ca-bound intermediate. Both conformations resemble their counterparts of the scrambling-incompetent anion channel mTMEM16A, yet with distinct differences in the region of ion and lipid permeation. In conjunction with functional data, we demonstrate the relationship between ion conduction and lipid scrambling. Although activated by a common mechanism, both functions appear to be mediated by alternate protein conformations that are at equilibrium in the ligand-bound state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4611.map.gz emd_4611.map.gz | 37.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4611-v30.xml emd-4611-v30.xml emd-4611.xml emd-4611.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4611_fsc.xml emd_4611_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_4611.png emd_4611.png | 197.1 KB | ||
| Masks |  emd_4611_msk_1.map emd_4611_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4611.cif.gz emd-4611.cif.gz | 6.8 KB | ||
| Others |  emd_4611_half_map_1.map.gz emd_4611_half_map_1.map.gz emd_4611_half_map_2.map.gz emd_4611_half_map_2.map.gz | 30.2 MB 30.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4611 http://ftp.pdbj.org/pub/emdb/structures/EMD-4611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4611 | HTTPS FTP |
-Related structure data
| Related structure data |  6qp6MC  4612C  4613C  4614C  6qpbC  6qpcC  6qpiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4611.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4611.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4611_msk_1.map emd_4611_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 used during refinement and for...
| File | emd_4611_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 used during refinement and for FSC gold-standard resolution calculation mTMEM16F_dig_Ca. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 used during refinement and for...
| File | emd_4611_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 used during refinement and for FSC gold-standard resolution calculation mTMEM16F_dig_Ca. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mTMEM16F
| Entire | Name: mTMEM16F |
|---|---|
| Components |
|
-Supramolecule #1: mTMEM16F
| Supramolecule | Name: mTMEM16F / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 212 KDa |
-Macromolecule #1: Anoctamin-6
| Macromolecule | Name: Anoctamin-6 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.367727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQMMTRKVLL NMELEEDDDE DGDIVLENFD QTIVCPTFGS LENQQDFRTP EFEEFNGKPD SLFFTDGQRR IDFILVYEDE SKKENNKKG TNEKQKRKRQ AYESNLICHG LQLEATRSVS DDKLVFVKVH APWEVLCTYA EIMHIKLPLK PNDLKTRSPF G NLNWFTKV ...String: MQMMTRKVLL NMELEEDDDE DGDIVLENFD QTIVCPTFGS LENQQDFRTP EFEEFNGKPD SLFFTDGQRR IDFILVYEDE SKKENNKKG TNEKQKRKRQ AYESNLICHG LQLEATRSVS DDKLVFVKVH APWEVLCTYA EIMHIKLPLK PNDLKTRSPF G NLNWFTKV LRVNESVIKP EQEFFTAPFE KSRMNDFYIL DRDSFFNPAT RSRIVYFILS RVKYQVMNNV NKFGINRLVS SG IYKAAFP LHDCRFNYES EDISCPSERY LLYREWAHPR SIYKKQPLDL IRKYYGEKIG IYFAWLGYYT QMLLLAAVVG VAC FLYGYL DQDNCTWSKE VCDPDIGGQI LMCPQCDRLC PFWRLNITCE SSKKLCIFDS FGTLIFAVFM GVWVTLFLEF WKRR QAELE YEWDTVELQQ EEQARPEYEA QCNHVVINEI TQEEERIPFT TCGKCIRVTL CASAVFFWIL LIIASVIGII VYRLS VFIV FSTTLPKNPN GTDPIQKYLT PQMATSITAS IISFIIIMIL NTIYEKVAIM ITNFELPRTQ TDYENSLTMK MFLFQF VNY YSSCFYIAFF KGKFVGYPGD PVYLLGKYRS EECDPGGCLL ELTTQLTIIM GGKAIWNNIQ EVLLPWVMNL IGRYKRV SG SEKITPRWEQ DYHLQPMGKL GLFYEYLEMI IQFGFVTLFV ASFPLAPLLA LVNNILEIRV DAWKLTTQFR RMVPEKAQ D IGAWQPIMQG IAILAVVTNA MIIAFTSDMI PRLVYYWSFS IPPYGDHTYY TMDGYINNTL SVFNITDFKN TDKENPYIG LGNYTLCRYR DFRNPPGHPQ EYKHNIYYWH VIAAKLAFII VMEHIIYSVK FFISYAIPDV SKITKSKIKR EKYLTQKLLH ESHLKDLTK NMGIIAERIG GTVDNSVRPK LE UniProtKB: Anoctamin-6 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: P1O |
|---|---|
| Molecular weight | Theoretical: 566.728 Da |
| Chemical component information | 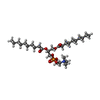 ChemComp-P1O: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 0.1% digitonin, 150 mM NaCl, 20 mM HEPES, pH 7.5 and 2 mM EGTA. 1 mM CaCl2 were added before freezing. |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: at 5 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 7 / Number real images: 5633 / Average exposure time: 9.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)