[English] 日本語
 Yorodumi
Yorodumi- EMDB-45530: STRUCTURE OF CD4 MIMETIC CJF-III-288 IN COMPLEX WITH BG505 SOSIP.... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF CD4 MIMETIC CJF-III-288 IN COMPLEX WITH BG505 SOSIP.664 HIV-1ENV TRIMER AND 17B FAB | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / FUSION / ENTRY / MEMBRANE / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Niu L / Tolbert WD / Pazgier M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC. Authors: Jonathan Richard / Michael W Grunst / Ling Niu / Marco A Díaz-Salinas / William D Tolbert / Lorie Marchitto / Fei Zhou / Catherine Bourassa / Derek Yang / Ta Jung Chiu / Hung-Ching Chen / ...Authors: Jonathan Richard / Michael W Grunst / Ling Niu / Marco A Díaz-Salinas / William D Tolbert / Lorie Marchitto / Fei Zhou / Catherine Bourassa / Derek Yang / Ta Jung Chiu / Hung-Ching Chen / Mehdi Benlarbi / / Suneetha Gottumukkala / Wenwei Li / Katrina Dionne / Étienne Bélanger / Debashree Chatterjee / Halima Medjahed / Wayne A Hendrickson / Joseph Sodroski / Zabrina C Lang / Abraham J Morton / Rick K Huang / Doreen Matthies / Amos B Smith / Walther Mothes / James B Munro / Marzena Pazgier / Andrés Finzi /   Abstract: HIV-1 envelope glycoproteins (Env) from primary HIV-1 isolates typically adopt a pretriggered "closed" conformation that resists to CD4-induced (CD4i) non-neutralizing antibodies (nnAbs) mediating ...HIV-1 envelope glycoproteins (Env) from primary HIV-1 isolates typically adopt a pretriggered "closed" conformation that resists to CD4-induced (CD4i) non-neutralizing antibodies (nnAbs) mediating antibody-dependent cellular cytotoxicity (ADCC). CD4-mimetic compounds (CD4mcs) "open-up" Env allowing binding of CD4i nnAbs, thereby sensitizing HIV-1-infected cells to ADCC. Two families of CD4i nnAbs, the anti-cluster A and anti-coreceptor binding site (CoRBS) Abs, are required to mediate ADCC in combination with the indane CD4mc BNM-III-170. Recently, new indoline CD4mcs with improved potency and breadth have been described. Here, we show that the lead indoline CD4mc, CJF-III-288, sensitizes HIV-1-infected cells to ADCC mediated by anti-CoRBS Abs alone, contributing to improved ADCC activity. Structural and conformational analyses reveal that CJF-III-288, in combination with anti-CoRBS Abs, potently stabilizes an asymmetric "open" State-3 Env conformation, This Env conformation orients the anti-CoRBS Ab to improve ADCC activity and therapeutic potential. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45530.map.gz emd_45530.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45530-v30.xml emd-45530-v30.xml emd-45530.xml emd-45530.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45530_fsc.xml emd_45530_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_45530.png emd_45530.png | 55.4 KB | ||
| Filedesc metadata |  emd-45530.cif.gz emd-45530.cif.gz | 7.9 KB | ||
| Others |  emd_45530_half_map_1.map.gz emd_45530_half_map_1.map.gz emd_45530_half_map_2.map.gz emd_45530_half_map_2.map.gz | 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45530 http://ftp.pdbj.org/pub/emdb/structures/EMD-45530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45530 | HTTPS FTP |
-Related structure data
| Related structure data |  9cf5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45530.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45530.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_45530_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_45530_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of BG505 SOSIP.664 Env trimer with CJF-III-288 and three ...
| Entire | Name: Complex of BG505 SOSIP.664 Env trimer with CJF-III-288 and three bound 17b FABS. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of BG505 SOSIP.664 Env trimer with CJF-III-288 and three ...
| Supramolecule | Name: Complex of BG505 SOSIP.664 Env trimer with CJF-III-288 and three bound 17b FABS. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: BG505 SOSIP.664 Env trimer
| Supramolecule | Name: BG505 SOSIP.664 Env trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: 17b Fab
| Supramolecule | Name: 17b Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Details: BG505 SOSIP.664 construct / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.864086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLWVTVYYGV PVWKDAETTL FCASDAKAYE TEKHNVWATH ACVPTDPNPQ EIHLENVTEE FNMWKNNMVE QMHTDIISLW DQSLKPCVK LTPLCVTLQC TNVTNNITDD MRGELKNCSF NMTTELRDKK QKVYSLFYRL DVVQINENQG NRSNNSNKEY R LINCNTSA ...String: NLWVTVYYGV PVWKDAETTL FCASDAKAYE TEKHNVWATH ACVPTDPNPQ EIHLENVTEE FNMWKNNMVE QMHTDIISLW DQSLKPCVK LTPLCVTLQC TNVTNNITDD MRGELKNCSF NMTTELRDKK QKVYSLFYRL DVVQINENQG NRSNNSNKEY R LINCNTSA ITQACPKVSF EPIPIHYCAP AGFAILKCKD KKFNGTGPCP SVSTVQCTHG IKPVVSTQLL LNGSLAEEEV MI RSENITN NAKNILVQFN TPVQINCTRP NNNTRKSIRI GPGQAFYATG DIIGDIRQAH CNVSKATWNE TLGKVVKQLR KHF GNNTII RFANSSGGDL EVTTHSFNCG GEFFYCNTSG LFNSTWISNT SVQGSNSTGS NDSITLPCRI KQIINMWQRI GQAM YAPPI QGVIRCVSNI TGLILTRDGG STNSTTETFR PGGGDMRDNW RSELYKYKVV KIEPLGVAPT RCKRRVVGRR RRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Details: BG505 SOSIP.664 construct / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: 17b Fab light chain
| Macromolecule | Name: 17b Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.399898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPAT LSVSPGERAT LSCRASESVS SDLAWYQQKP GQAPRLLIYG ASTRATGVPA RFSGSGSGAE FTLTISSLQS EDFAVYYCQ QYNNWPPRYT FGQGTRLEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: DIVMTQSPAT LSVSPGERAT LSCRASESVS SDLAWYQQKP GQAPRLLIYG ASTRATGVPA RFSGSGSGAE FTLTISSLQS EDFAVYYCQ QYNNWPPRYT FGQGTRLEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRG |
-Macromolecule #4: 17b Fab heavy chain
| Macromolecule | Name: 17b Fab heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.572479 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGAE VKKPGSSVKV SCKASGDTFI RYSFTWVRQA PGQGLEWMGR IITILDVAHY APHLQGRVTI TADKSTSTVY LELRNLRSD DTAVYFCAGV YEGEADEGEY DNNGFLKHWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS ...String: EVQLVESGAE VKKPGSSVKV SCKASGDTFI RYSFTWVRQA PGQGLEWMGR IITILDVAHY APHLQGRVTI TADKSTSTVY LELRNLRSD DTAVYFCAGV YEGEADEGEY DNNGFLKHWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKRVEP KS |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 26 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: propyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroa...
| Macromolecule | Name: propyl (2R,3S)-2-(carbamimidamidomethyl)-3-[2-(4-chloro-3-fluoroanilino)(oxo)acetamido]-6-[(methylamino)methyl]-2,3-dihydro-1H-indole-1-carboxylate type: ligand / ID: 9 / Number of copies: 3 / Formula: Y26 |
|---|---|
| Molecular weight | Theoretical: 533.983 Da |
| Chemical component information | 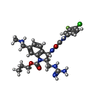 ChemComp-Y26: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9431 / Average electron dose: 54.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































