[English] 日本語
 Yorodumi
Yorodumi- EMDB-45217: Human DNA polymerase theta helicase domain in microhomology annea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human DNA polymerase theta helicase domain in microhomology annealed state 1, dimer form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair / helicase / ATPase / TRANSFERASE-DNA complex / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
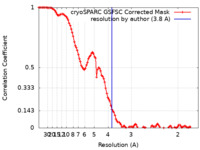
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Ito F / Li Z / Chen XS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2024 Title: Structural Basis for Polθ-Helicase DNA Binding and Microhomology-Mediated End-Joining. Authors: Fumiaki Ito / Ziyuan Li / Leonid Minakhin / Htet A Khant / Richard T Pomerantz / Xiaojiang S Chen /  Abstract: DNA double-strand breaks (DSBs) present a critical threat to genomic integrity, often precipitating genomic instability and oncogenesis. Repair of DSBs predominantly occurs through homologous ...DNA double-strand breaks (DSBs) present a critical threat to genomic integrity, often precipitating genomic instability and oncogenesis. Repair of DSBs predominantly occurs through homologous recombination (HR) and non-homologous end joining (NHEJ). In HR-deficient cells, DNA polymerase theta (Polθ) becomes critical for DSB repair via microhomology-mediated end joining (MMEJ), also termed theta-mediated end joining (TMEJ). Thus, Polθ is synthetically lethal with BRCA1/2 and other HR factors, underscoring its potential as a therapeutic target in HR-deficient cancers. However, the molecular mechanisms governing Polθ-mediated MMEJ remain poorly understood. Here we present a series of cryo-electron microscopy structures of the Polθ helicase domain (Polθ-hel) in complex with DNA containing 3'-overhang. The structures reveal the sequential conformations adopted by Polθ-hel during the critical phases of DNA binding, microhomology searching, and microhomology annealing. The stepwise conformational changes within the Polθ-hel subdomains and its functional dimeric state are pivotal for aligning the 3'-overhangs, facilitating the microhomology search and subsequent annealing necessary for DSB repair via MMEJ. Our findings illustrate the essential molecular switches within Polθ-hel that orchestrate the MMEJ process in DSB repair, laying the groundwork for the development of targeted therapies against the Polθ-hel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45217.map.gz emd_45217.map.gz | 63.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45217-v30.xml emd-45217-v30.xml emd-45217.xml emd-45217.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45217_fsc.xml emd_45217_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_45217.png emd_45217.png | 122.8 KB | ||
| Filedesc metadata |  emd-45217.cif.gz emd-45217.cif.gz | 4.2 KB | ||
| Others |  emd_45217_half_map_1.map.gz emd_45217_half_map_1.map.gz emd_45217_half_map_2.map.gz emd_45217_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45217 http://ftp.pdbj.org/pub/emdb/structures/EMD-45217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45217 | HTTPS FTP |
-Validation report
| Summary document |  emd_45217_validation.pdf.gz emd_45217_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45217_full_validation.pdf.gz emd_45217_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_45217_validation.xml.gz emd_45217_validation.xml.gz | 18.8 KB | Display | |
| Data in CIF |  emd_45217_validation.cif.gz emd_45217_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45217 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45217 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45217 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45217 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45217.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45217.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_45217_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
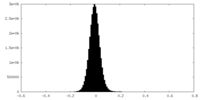
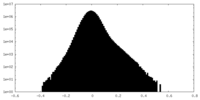
| Density Histograms |
-Half map: #2
| File | emd_45217_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
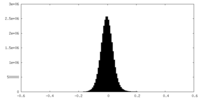
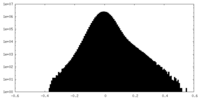
| Density Histograms |
- Sample components
Sample components
-Entire : Human DNA polymerase theta helicase domain in microhomology annea...
| Entire | Name: Human DNA polymerase theta helicase domain in microhomology annealed state 1, dimer form |
|---|---|
| Components |
|
-Supramolecule #1: Human DNA polymerase theta helicase domain in microhomology annea...
| Supramolecule | Name: Human DNA polymerase theta helicase domain in microhomology annealed state 1, dimer form type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 253 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 10331 / Average exposure time: 3.5 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)