[English] 日本語
 Yorodumi
Yorodumi- EMDB-45214: Structure of Human Adaptor Protein Complex AP-3 in the Apo State -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Human Adaptor Protein Complex AP-3 in the Apo State | |||||||||
 Map data Map data | deepEMHancer map of human AP-3 in the Apo state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adaptor Protein complex / AP-3 / Lysosomal transport / Endosomal transport / Protein trafficking / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsynaptic vesicle coating / synaptic vesicle budding from endosome / : / clathrin-coated vesicle cargo loading, AP-3-mediated / skin epidermis development / AP-type membrane coat adaptor complex / synaptic vesicle membrane organization / zinc ion import into lysosome / AP-3 adaptor complex / neurotransmitter receptor transport, postsynaptic endosome to lysosome ...synaptic vesicle coating / synaptic vesicle budding from endosome / : / clathrin-coated vesicle cargo loading, AP-3-mediated / skin epidermis development / AP-type membrane coat adaptor complex / synaptic vesicle membrane organization / zinc ion import into lysosome / AP-3 adaptor complex / neurotransmitter receptor transport, postsynaptic endosome to lysosome / anterograde synaptic vesicle transport / microvesicle / endosome to melanosome transport / synaptic vesicle recycling / postsynaptic recycling endosome / Golgi to vacuole transport / clathrin adaptor complex / platelet dense granule organization / melanosome assembly / presynaptic endosome / postsynaptic neurotransmitter receptor internalization / granulocyte differentiation / positive regulation of NK T cell differentiation / clathrin-coated vesicle membrane / antigen processing and presentation, exogenous lipid antigen via MHC class Ib / protein targeting to vacuole / GTP-dependent protein binding / melanosome organization / protein targeting to lysosome / respiratory system process / anterograde axonal transport / intracellular zinc ion homeostasis / protein localization to cell surface / protein localization to membrane / lysosome organization / toll-like receptor signaling pathway / Golgi Associated Vesicle Biogenesis / Association of TriC/CCT with target proteins during biosynthesis / lung morphogenesis / homeostasis of number of cells / single fertilization / intracellular transport / hematopoietic progenitor cell differentiation / transport vesicle / vesicle-mediated transport / axon cytoplasm / cytoplasmic vesicle membrane / intracellular protein transport / mRNA transcription by RNA polymerase II / protein modification process / small GTPase binding / endocytosis / cell morphogenesis / terminal bouton / blood coagulation / Signaling by BRAF and RAF1 fusions / insulin receptor signaling pathway / presynapse / cytoplasmic vesicle / protein phosphatase binding / spermatogenesis / early endosome / lysosome / postsynapse / endosome membrane / inflammatory response / Golgi membrane / lysosomal membrane / intracellular membrane-bounded organelle / glutamatergic synapse / Golgi apparatus / positive regulation of transcription by RNA polymerase II / mitochondrion / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
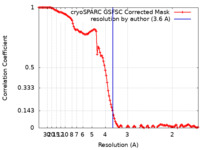
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Begley MC / Baker RW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: A structure-based mechanism for initiation of AP-3 coated vesicle formation. Authors: Matthew Begley / Mahira Aragon / Richard W Baker /  Abstract: Adaptor protein complex-3 (AP-3) mediates cargo sorting from endosomes to lysosomes and lysosome-related organelles. Recently, it was shown that AP-3 adopts a constitutively open conformation ...Adaptor protein complex-3 (AP-3) mediates cargo sorting from endosomes to lysosomes and lysosome-related organelles. Recently, it was shown that AP-3 adopts a constitutively open conformation compared to the related AP-1 and AP-2 coat complexes, which are inactive until undergoing large conformational changes upon membrane recruitment. How AP-3 is regulated is therefore an open question. To understand the mechanism of AP-3 membrane recruitment and activation, we reconstituted human AP-3 and determined multiple structures in the soluble and membrane-bound states using electron cryo-microscopy. Similar to yeast AP-3, human AP-3 is in a constitutively open conformation. To reconstitute AP-3 activation by adenosine di-phosphate (ADP)-ribosylation factor 1 (Arf1), a small guanosine tri-phosphate (GTP)ase, we used lipid nanodiscs to build Arf1-AP-3 complexes on membranes and determined three structures showing the stepwise conformational changes required for formation of AP-3 coated vesicles. First, membrane recruitment is driven by one of two predicted Arf1 binding sites, which flexibly tethers AP-3 to the membrane. Second, cargo binding causes AP-3 to adopt a fixed position and rigidifies the complex, which stabilizes binding for a second Arf1 molecule. Finally, binding of the second Arf1 molecule provides the template for AP-3 dimerization, providing a glimpse into the first step of coat polymerization. We propose coat polymerization only occurs after cargo engagement, thereby linking cargo sorting with assembly of higher-order coat structures. Additionally, we provide evidence for two amphipathic helices in AP-3, suggesting that AP-3 contributes to membrane deformation during coat assembly. In total, these data provide evidence for the first stages of AP-3-mediated vesicle coat assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45214.map.gz emd_45214.map.gz | 158 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45214-v30.xml emd-45214-v30.xml emd-45214.xml emd-45214.xml | 40.9 KB 40.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45214_fsc.xml emd_45214_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_45214.png emd_45214.png | 93.8 KB | ||
| Masks |  emd_45214_msk_1.map emd_45214_msk_1.map emd_45214_msk_2.map emd_45214_msk_2.map | 178 MB 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45214.cif.gz emd-45214.cif.gz | 8.6 KB | ||
| Others |  emd_45214_additional_1.map.gz emd_45214_additional_1.map.gz emd_45214_additional_2.map.gz emd_45214_additional_2.map.gz emd_45214_additional_3.map.gz emd_45214_additional_3.map.gz emd_45214_additional_4.map.gz emd_45214_additional_4.map.gz emd_45214_additional_5.map.gz emd_45214_additional_5.map.gz emd_45214_half_map_1.map.gz emd_45214_half_map_1.map.gz emd_45214_half_map_2.map.gz emd_45214_half_map_2.map.gz | 4.4 MB 4.1 MB 156.3 MB 167.7 MB 87.6 MB 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45214 http://ftp.pdbj.org/pub/emdb/structures/EMD-45214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45214 | HTTPS FTP |
-Validation report
| Summary document |  emd_45214_validation.pdf.gz emd_45214_validation.pdf.gz | 685.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45214_full_validation.pdf.gz emd_45214_full_validation.pdf.gz | 685.3 KB | Display | |
| Data in XML |  emd_45214_validation.xml.gz emd_45214_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_45214_validation.cif.gz emd_45214_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45214 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45214 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45214 | HTTPS FTP |
-Related structure data
| Related structure data |  9c5cMC  9c58C  9c59C  9c5aC  9c5bC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45214.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45214.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMHancer map of human AP-3 in the Apo state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45214_msk_1.map emd_45214_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_45214_msk_2.map emd_45214_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map filter by local resolution for human AP-3 in the Apo state
| File | emd_45214_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map filter by local resolution for human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map for coloring by local resolution
| File | emd_45214_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map for coloring by local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor sharpened map of human AP-3 in the Apo state
| File | emd_45214_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map of human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor sharpened map of human AP-3 in the Apo state
| File | emd_45214_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map of human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Filtered map of human AP-3 in the Apo state
| File | emd_45214_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered map of human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map of human AP-3 in the Apo state
| File | emd_45214_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map of human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map of human AP-3 in the Apo state
| File | emd_45214_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map of human AP-3 in the Apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Adaptor Protein Complex AP-3 in the Apo state
| Entire | Name: Human Adaptor Protein Complex AP-3 in the Apo state |
|---|---|
| Components |
|
-Supramolecule #1: Human Adaptor Protein Complex AP-3 in the Apo state
| Supramolecule | Name: Human Adaptor Protein Complex AP-3 in the Apo state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 215 KDa |
-Macromolecule #1: AP-3 complex subunit delta-1
| Macromolecule | Name: AP-3 complex subunit delta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.164953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LQDLVRGIRN HKEDEAKYIS QCIDEIKQEL KQDNIAVKAN AVCKLTYLQM LGYDISWAAF NIIEVMSASK FTFKRIGYLA ASQSFHEGT DVIMLTTNQI RKDLSSPSQY DTGVALTGLS CFVTPDLARD LANDIMTLMS HTKPYIRKKA VLIMYKVFLK Y PESLRPAF ...String: LQDLVRGIRN HKEDEAKYIS QCIDEIKQEL KQDNIAVKAN AVCKLTYLQM LGYDISWAAF NIIEVMSASK FTFKRIGYLA ASQSFHEGT DVIMLTTNQI RKDLSSPSQY DTGVALTGLS CFVTPDLARD LANDIMTLMS HTKPYIRKKA VLIMYKVFLK Y PESLRPAF PRLKEKLEDP DPGVQSAAVN VICELARRNP KNYLSLAPLF FKLMTSSTNN WVLIKIIKLF GALTPLEPRL GK KLIEPLT NLIHSTSAMS LLYECVNTVI AVLISLSSGM PNHSASIQLC VQKLRILIED SDQNLKYLGL LAMSKILKTH PKS VQSHKD LILQCLDDKD ESIRLRALDL LYGMVSKKNL MEIVKKLMTH VDKAEGTTYR DELLTKIIDI CSQSNYQYIT NFEW YISIL VELTRLEGTR HGHLIAAQML DVAIRVKAIR KFAVSQMSAL LDSAHLLASS TQRNGICEVL YAAAWICGEF SEHLQ EPHH TLEAMLRPRV TTLPGHIQAV YVQNVVKLYA SILQQKEQAG EAEGAQAVTQ LMVDRLPQFV QSADLEVQER ASCILQ LVK HIQKLQAKDV PVAEEVSALF AGELN UniProtKB: AP-3 complex subunit delta-1 |
-Macromolecule #2: AP-3 complex subunit beta-1
| Macromolecule | Name: AP-3 complex subunit beta-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.602797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DLKKNEDLKQ MLESNKDSAK LDAMKRIVGM IAKGKNASEL FPAVVKNVAS KNIEIKKLVY VYLVRYAEEQ QDLALLSIST FQRALKDPN QLIRASALRV LSSIRVPIIV PIMMLAIKEA SADLSPYVRK NAAHAIQKLY SLDPEQKEML IEVIEKLLKD K STLVAGSV ...String: DLKKNEDLKQ MLESNKDSAK LDAMKRIVGM IAKGKNASEL FPAVVKNVAS KNIEIKKLVY VYLVRYAEEQ QDLALLSIST FQRALKDPN QLIRASALRV LSSIRVPIIV PIMMLAIKEA SADLSPYVRK NAAHAIQKLY SLDPEQKEML IEVIEKLLKD K STLVAGSV VMAFEEVCPD RIDLIHKNYR KLCNLLVDVE EWGQVVIIHM LTRYARTQFV SPWDPDHRLL IRNTKPLLQS RN AAVVMAV AQLYWHISPK SEAGIISKSL VRLLRSNREV QYIVLQNIAT MSIQRKGMFE PYLKSFYVRS TDPTMIKTLK LEI LTNLAN EANISTLLRE FQTYVKSQDK QFAAATIQTI GRCATNILEV TDTCLNGLVC LLSNRDEIVV AESVVVIKKL LQMQ PAQHG EIIKHMAKLL DSITVPVARA SILWLIGENC ERVPKIAPDV LRKMAKSFTS EDDLVKLQIL NLGAKLYLTN SKQTK LLTQ YILNLGKYDQ NYDIRDRTRF IRQLIVPNVK SGALSKYAKK IFLAQKPAPL LESPFKDRDH FQLGTLSHTL NIKATG YLE LSNWPEVAPD PSVRN UniProtKB: AP-3 complex subunit beta-1 |
-Macromolecule #3: AP-3 complex subunit mu-1
| Macromolecule | Name: AP-3 complex subunit mu-1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.274275 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIHSLFLINC SGDIFLEKHW KSVVSQSVCD YFFEAQEKAA DVENVPPVIS TPHHYLISIY RDKLFFVSVI QTEVPPLFVI EFLHRVADT FQDYFGECSE AAIKDNVVIV YELLEEMLDN GFPLA UniProtKB: AP-3 complex subunit mu-1 |
-Macromolecule #4: AP-3 complex subunit sigma-1
| Macromolecule | Name: AP-3 complex subunit sigma-1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.412949 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKAILIFNN HGKPRLSKFY QPYSEDTQQQ IIRETFHLVS KRDENVCNFL EGGLLIGGSD NKLIYRHYAT LYFVFCVDSS ESELGILDL IQVFVETLDK CFENVCELDL IFHVDKVHNI LAEMVMGGMV LETNMNEIVT QIDAQNKLEK SE UniProtKB: AP-3 complex subunit sigma-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 1x PBS (pH 7.4), 300mM NaCl, 1mM TCEP | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR Details: Used Quantifoil Active grids- Backside gold coated before plasma cleaned. 12 mA used for plasma cleaning | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 296 K / Instrument: SPOTITON / Details: Commercialized version - Chameleon. | ||||||||||||||||||
| Details | Specimen appeared as a monodisperse peak via size exclusion chromatography (SEC) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 49.97 e/Å2 Details: 4 datasets collected, processed independently, and merged. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-9c5c: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)