[English] 日本語
 Yorodumi
Yorodumi- EMDB-45196: Cryo-EM structure of the Strand displacement Complex (III) of Yea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Strand displacement Complex (III) of Yeast Mitochondrial DNA polymerase Gamma (MIP1) with downstream DNA | |||||||||
 Map data Map data | Anisotropic sharpened map of the original map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondrial DNA Polymerase Gamma / Strand displacement complex / MIP1 / REPLICATION / Helicase independent DNA polymerase / Transferase-DNA complex | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Nayak AR / Sokolova VO / Sillamaa S / Sedmen J / Temiakov D / Zamudio-Ochoa A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural basis for intrinsic strand displacement activity of mitochondrial DNA polymerase. Authors: Ashok R Nayak / Viktoriia Sokolova / Sirelin Sillamaa / Karl Herbine / Juhan Sedman / Dmitry Temiakov /   Abstract: Members of the Pol A family of DNA polymerases, found across all domains of life, utilize various strategies for DNA strand separation during replication. In higher eukaryotes, mitochondrial DNA ...Members of the Pol A family of DNA polymerases, found across all domains of life, utilize various strategies for DNA strand separation during replication. In higher eukaryotes, mitochondrial DNA polymerase γ relies on the replicative helicase TWINKLE, whereas the yeast ortholog, Mip1, can unwind DNA independently. Using Mip1 as a model, we present a series of high-resolution cryo-EM structures that capture the process of DNA strand displacement. Our data reveal previously unidentified structural elements that facilitate the unwinding of the downstream DNA duplex. Yeast cells harboring Mip1 variants defective in strand displacement exhibit impaired oxidative phosphorylation and loss of mtDNA, corroborating the structural observations. This study provides a molecular basis for the intrinsic strand displacement activity of Mip1 and illuminates the distinct unwinding mechanisms utilized by Pol A family DNA polymerases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45196.map.gz emd_45196.map.gz | 134.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45196-v30.xml emd-45196-v30.xml emd-45196.xml emd-45196.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45196_fsc.xml emd_45196_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_45196.png emd_45196.png | 115.4 KB | ||
| Filedesc metadata |  emd-45196.cif.gz emd-45196.cif.gz | 8.4 KB | ||
| Others |  emd_45196_additional_1.map.gz emd_45196_additional_1.map.gz emd_45196_additional_2.map.gz emd_45196_additional_2.map.gz emd_45196_half_map_1.map.gz emd_45196_half_map_1.map.gz emd_45196_half_map_2.map.gz emd_45196_half_map_2.map.gz | 126 MB 72.7 MB 134.3 MB 134.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45196 http://ftp.pdbj.org/pub/emdb/structures/EMD-45196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45196 | HTTPS FTP |
-Related structure data
| Related structure data |  9c53MC  9c51C  9c52C  9d2iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45196.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45196.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anisotropic sharpened map of the original map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7653 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEM sharpened map
| File | emd_45196_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEM sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Original unsharpened map
| File | emd_45196_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_45196_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_45196_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM map of the Strand displacement Complex (III) of Yeast Mit...
| Entire | Name: Cryo-EM map of the Strand displacement Complex (III) of Yeast Mitochondrial DNA polymerase Gamma (MIP1) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of the Strand displacement Complex (III) of Yeast Mit...
| Supramolecule | Name: Cryo-EM map of the Strand displacement Complex (III) of Yeast Mitochondrial DNA polymerase Gamma (MIP1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Yeast Mitochondrial DNA polymerase Gamma (MIP1)wild-type assembled on a primer-template and a downstream non-template strand with incoming nucleotide, dATP bound to the polymerase site. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142 KDa |
-Macromolecule #1: DNA polymerase gamma
| Macromolecule | Name: DNA polymerase gamma / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.745438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASTKKNTAE APRINPVGIQ YLGESLQRQV FGSCGGKDEV EQSDKLMELS KKSLKDHGLW GKKTLITDPI SFPLPPLQGR SLDEHFQKI GRFNSEPYKS FCEDKFTEMV ARPAEWLRKP GWVKYVPGMA PVEVAYPDEE LVVFDVETLY NVSDYPTLAT A LSSTAWYL ...String: MASTKKNTAE APRINPVGIQ YLGESLQRQV FGSCGGKDEV EQSDKLMELS KKSLKDHGLW GKKTLITDPI SFPLPPLQGR SLDEHFQKI GRFNSEPYKS FCEDKFTEMV ARPAEWLRKP GWVKYVPGMA PVEVAYPDEE LVVFDVETLY NVSDYPTLAT A LSSTAWYL WCSPFICGGD DPAALIPLNT LNKEQVVIGH NVAYDRARVL EEYNFRDSKA FFLDTQSLHI ASFGLCSRQR PM FMKNNKK KEAEVESEVH PEISIEDYDD PWLNVSALNS LKDVAKFHCK IDLDKTDRDF FASTDKSTII ENFQKLVNYC ATD VTATSQ VFDKIFPVFL KKCPHPVSFA GLKSLSKCIL PTKLNDWNDY LNSSESLYQQ SKVQIESKIV QIIKDIALLK DKPD FYLKD PWLSQLDWTT KPLRLTKKGV PAKCQKLPGF PEWYRQLFPS KDTVEPKITI KSRIIPILFK LSWENSPVIW SKESG WCFN VPHEQVETYK AKNYVLADSV SQEEEEIRMN NLGLQCTGVL FKVPHPNGPT FNCTNLLTKS YNHFFEKGVL KSESEL AHQ ALQINSSGSY WMSARERIQS QFVVPNCKFP NEFQSLSAKS SLNNEKTNDL AIIIPKIVPM GTITRRAVEN TWLTASN AK ANRIGSELKT QVKAPPGYCF VGADVDSEEL WIASLVGDSI FNVHGGTAIG WMCLEGTKNE GTDLHTKTAQ ILGCSRNE A KIFNYGRIYG AGAKFASQLL KRFNPSLTDE ETKKIANKLY ENTKGKTKRS KLFKKFWYGG SESILFNKLE SIAEQETPK TPVLGCGITY SLMKKNLRAN SFLPSRINWA IQSSGVDYLH LLCCSMEYII KKYNLEARLC ISIHDEIRFL VSEKDKYRAA MALQISNIW TRAMFCQQMG INELPQNCAF FSQVDIDSVI RKEVNMDCIT PSNKTAIPHG EALDINQLLD KPNSKLGKPS L DIDSKVSQ YAYNYREPVF EEYNKSYTPE FLKYFLAMQV QSDKRDVNRL EDEYLRECTS KEYARDGNTA EYSLLDYIKD VE KGKRTKV RIMGSNFLDG TKNAKADQRI RLPVNMPDYP TLHKIANDSA IPEKQLLENR RKKENRIDDE NKKKLTRKKN TTP MERKYK RVYGGRKAFE AFYECANKPL DYTLETEKQF FNIPIDGVID DVLNDKSNYK KKPSQARTAS SSPIRKTAKA VHSK KLPAR KSSTTNRNLV ELERDITISR EYKLAAALEH HHHHH UniProtKB: UNIPROTKB: A0A8H4BW69 |
-Macromolecule #2: Non-Template DNA
| Macromolecule | Name: Non-Template DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.078555 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DG)(DG)(DT)(DA)(DG)(DT) (DC)(DG)(DT)(DG)(DA)(DC)(DT)(DC)(DG)(DA) (DC)(DC)(DG) |
-Macromolecule #3: Primer DNA
| Macromolecule | Name: Primer DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.17261 KDa |
| Sequence | String: (DG)(DA)(DA)(DG)(DA)(DC)(DA)(DG)(DT)(DC) (DT)(DG)(DC)(DG)(DG)(DC)(DG)(DC)(DG)(DC) (DG)(DG)(DG) |
-Macromolecule #4: Template DNA
| Macromolecule | Name: Template DNA / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.854218 KDa |
| Sequence | String: (DC)(DG)(DG)(DT)(DC)(DG)(DA)(DG)(DA)(DG) (DT)(DC)(DA)(DC)(DG)(DA)(DC)(DT)(DA)(DC) (DC)(DC)(DG)(DC)(DG)(DC)(DG)(DC)(DC) (DG)(DC)(DA)(DG)(DA)(DC)(DT)(DG)(DT)(DC) (DT) (DT)(DC) |
-Macromolecule #5: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: DTP |
|---|---|
| Molecular weight | Theoretical: 491.182 Da |
| Chemical component information |  ChemComp-DTP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: 3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE
| Macromolecule | Name: 3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: 2DT |
|---|---|
| Molecular weight | Theoretical: 306.209 Da |
| Chemical component information | 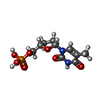 ChemComp-2DT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.57 mg/mL |
|---|---|
| Buffer | pH: 7.9 / Details: 10 mM Tris-HCL, 100 mM NaCL, 10 mM DTT, 5 mM MgCl2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | A 4 uM strand displacement complex complex was assembled with wild-type MIP1 and a DNA scaffold with primer, template, and non-template strands, mixed in equal millimolar ratio. The primer strand was extended by three nucleotides with 1 mM dGTP and chain terminated with 1 mM di-deoxy TTP. 1 mM dATP was added before plunge freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 10 eV |
| Software | Name: EPU (ver. 3.7) |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 2 / Number real images: 14819 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Residue range: 1-1254 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Software | Name:  Coot (ver. 0.9.8.5) Coot (ver. 0.9.8.5) |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9c53: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)






















































