[English] 日本語
 Yorodumi
Yorodumi- EMDB-45081: Phosphorylated human NKCC1_K289NA492E in complex with furosemide -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phosphorylated human NKCC1_K289NA492E in complex with furosemide | |||||||||
 Map data Map data | Phosphorylated human NKCC1_K289NA492E in complex with furosemide | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sodium potassium chloride cotransporter / phosphorylation / outward-open state / furosemide / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / metal ion transmembrane transporter activity / cell body membrane / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity ...positive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / metal ion transmembrane transporter activity / cell body membrane / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / transepithelial chloride transport / potassium ion transmembrane transporter activity / intracellular chloride ion homeostasis / negative regulation of vascular wound healing / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / cell projection membrane / cellular response to chemokine / T cell chemotaxis / potassium ion homeostasis / intracellular sodium ion homeostasis / sodium ion import across plasma membrane / cellular response to potassium ion / cell volume homeostasis / hyperosmotic response / regulation of spontaneous synaptic transmission / gamma-aminobutyric acid signaling pathway / maintenance of blood-brain barrier / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / lateral plasma membrane / transport across blood-brain barrier / monoatomic ion transport / chloride transmembrane transport / basal plasma membrane / cytoplasmic vesicle membrane / cell projection / sodium ion transmembrane transport / cell periphery / Hsp90 protein binding / protein-folding chaperone binding / extracellular vesicle / cell body / basolateral plasma membrane / apical plasma membrane / neuron projection / neuronal cell body / protein kinase binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Zhao YX / Cao EH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2025 Journal: EMBO J / Year: 2025Title: Structural basis for human NKCC1 inhibition by loop diuretic drugs. Authors: Yongxiang Zhao / Pietro Vidossich / Biff Forbush / Junfeng Ma / Jesse Rinehart / Marco De Vivo / Erhu Cao /    Abstract: Na-K-Cl cotransporters functions as an anion importers, regulating trans-epithelial chloride secretion, cell volume, and renal salt reabsorption. Loop diuretics, including furosemide, bumetanide, and ...Na-K-Cl cotransporters functions as an anion importers, regulating trans-epithelial chloride secretion, cell volume, and renal salt reabsorption. Loop diuretics, including furosemide, bumetanide, and torsemide, antagonize both NKCC1 and NKCC2, and are first-line medicines for the treatment of edema and hypertension. NKCC1 activation by the molecular crowding sensing WNK kinases is critical if cells are to combat shrinkage during hypertonic stress; however, how phosphorylation accelerates NKCC1 ion transport remains unclear. Here, we present co-structures of phospho-activated NKCC1 bound with furosemide, bumetanide, or torsemide showing that furosemide and bumetanide utilize a carboxyl group to coordinate and co-occlude a K, whereas torsemide encroaches and expels the K from the site. We also found that an amino-terminal segment of NKCC1, once phosphorylated, interacts with the carboxyl-terminal domain, and together, they engage with intracellular ion exit and appear to be poised to facilitate rapid ion translocation. Together, these findings enhance our understanding of NKCC-mediated epithelial ion transport and the molecular mechanisms of its inhibition by loop diuretics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45081.map.gz emd_45081.map.gz | 162.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45081-v30.xml emd-45081-v30.xml emd-45081.xml emd-45081.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_45081.png emd_45081.png | 50.2 KB | ||
| Filedesc metadata |  emd-45081.cif.gz emd-45081.cif.gz | 7.1 KB | ||
| Others |  emd_45081_additional_1.map.gz emd_45081_additional_1.map.gz emd_45081_half_map_1.map.gz emd_45081_half_map_1.map.gz emd_45081_half_map_2.map.gz emd_45081_half_map_2.map.gz | 164.5 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45081 http://ftp.pdbj.org/pub/emdb/structures/EMD-45081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45081 | HTTPS FTP |
-Related structure data
| Related structure data |  9c0eMC  9c0gC  9c0hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45081.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45081.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phosphorylated human NKCC1_K289NA492E in complex with furosemide | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8256 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Additional Map
| File | emd_45081_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_45081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_45081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phosphorylated NKCC1_K289NA492E in complex with furosemide
| Entire | Name: Phosphorylated NKCC1_K289NA492E in complex with furosemide |
|---|---|
| Components |
|
-Supramolecule #1: Phosphorylated NKCC1_K289NA492E in complex with furosemide
| Supramolecule | Name: Phosphorylated NKCC1_K289NA492E in complex with furosemide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 2
| Macromolecule | Name: Solute carrier family 12 member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 131.866328 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEPRPTAPSS GAPGLAGVGE TPSAAALAAA RVELPGTAVP SVPEDAAPAS RDGGGVRDEG PAAAGDGLGR PLGPTPSQSR FQVDLVSEN AGRAAAAAAA AAAAAAAAGA GAGAKQTPAD GEASGESEPA KGSEEAKGRF RVNFVDPAAS SSAEDSLSDA A GVGVDGPN ...String: MEPRPTAPSS GAPGLAGVGE TPSAAALAAA RVELPGTAVP SVPEDAAPAS RDGGGVRDEG PAAAGDGLGR PLGPTPSQSR FQVDLVSEN AGRAAAAAAA AAAAAAAAGA GAGAKQTPAD GEASGESEPA KGSEEAKGRF RVNFVDPAAS SSAEDSLSDA A GVGVDGPN VSFQNGGDTV LSEGSSLHSG GGGGSGHHQH YYYDTHTNTY YLR(TPO)FGHN(TPO)M DAVPRIDHYR H (TPO)AAQLGEK LLRPSLAELH DELEKEPFED GFANGEESTP TRDAVVTYTA ESKGVVKFGW INGVLVRCML NIWGVMLF I RLSWIVGQAG IGLSVLVIMM ATVVTTITGL STSAIATNGF VRGGGAYYLI SRSLGPEFGG AIGLIFAFAN AVAVAMYVV GFAETVVELL KEHSILMIDE INDIRIIGAI TVVILLGISV AGMEWEAKAQ IVLLVILLLA IGDFVIGTFI PLESKKPKGF FGYKSEIFN ENFGPDFREE ETFFSVFEIF FPAATGILAG ANISGDLADP QSAIPKGTLL AILITTLVYV GIAVSVGSCV V RDATGNVN DTIVTELTNC TSAACKLNFD FSSCESSPCS YGLMNNFQVM SMVSGFTPLI SAGIFSATLS SALASLVSAP KI FQALCKD NIYPAFQMFA KGYGKNNEPL RGYILTFLIA LGFILIAELN VIAPIISNFF LASYALINFS VFHASLAKSP GWR PAFKYY NMWISLLGAI LCCIVMFVIN WWAALLTYVI VLGLYIYVTY KKPDVNWGSS TQALTYLNAL QHSIRLSGVE DHVK NFRPQ CLVMTGAPNS RPALLHLVHD FTKNVGLMIC GHVHMGPRRQ AMKEMSIDQA KYQRWLIKNK MKAFYAPVHA DDLRE GAQY LMQAAGLGRM KPNTLVLGFK KDWLQADMRD VDMYINLFHD AFDIQYGVVV IRLKEGLDIS HLQGQEELLS SQEKSP GTK DVVVSVEYSK KSDLDTSKPL SEKPITHKVE EEDGKTATQP LLKKESKGPI VPLNVADQKL LEASTQFQKK QGKNTID VW WLFDDGGLTL LIPYLLTTKK KWKDCKIRVF IGGKINRIDH DRRAMATLLS KFRIDFSDIM VLGDINTKPK KENIIAFE E IIEPYRLHED DKEQDIADKM KEDEPWRITD NELELYKTKT YRQIRLNELL KEHSSTANII VMSLPVARKG AVSSALYMA WLEALSKDLP PILLVRGNHQ SVLTFYS UniProtKB: Solute carrier family 12 member 2 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID
| Macromolecule | Name: 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID type: ligand / ID: 3 / Number of copies: 2 / Formula: FUN |
|---|---|
| Molecular weight | Theoretical: 330.744 Da |
| Chemical component information | 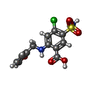 ChemComp-FUN: |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI MORGAGNI |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7000000000000001 µm |
 Movie
Movie Controller
Controller





 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)