[English] 日本語
 Yorodumi
Yorodumi- EMDB-44964: Homomeric alpha3 glycine receptor in the presence of 0.1 mM glyci... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Homomeric alpha3 glycine receptor in the presence of 0.1 mM glycine at pH 6.4 in an apo state | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Glycine / Ion Channel / Ligand-Gated / Pentameric / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglycine-gated chloride ion channel activity / glycine-gated chloride channel complex / Neurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated chloride channel activity / glycinergic synapse / glycine binding / presynaptic modulation of chemical synaptic transmission / chloride transmembrane transport / protein homooligomerization / transmembrane signaling receptor activity ...glycine-gated chloride ion channel activity / glycine-gated chloride channel complex / Neurotransmitter receptors and postsynaptic signal transmission / extracellularly glycine-gated chloride channel activity / glycinergic synapse / glycine binding / presynaptic modulation of chemical synaptic transmission / chloride transmembrane transport / protein homooligomerization / transmembrane signaling receptor activity / presynaptic membrane / perikaryon / postsynaptic membrane / intracellular membrane-bounded organelle / dendrite / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
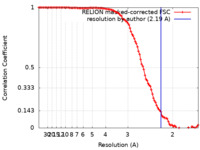
| Method | single particle reconstruction / cryo EM / Resolution: 2.19 Å | |||||||||||||||||||||
 Authors Authors | Kindig K / Gibbs E / Chakrapani S | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Mechanisms underlying modulation of human GlyRα3 by Zn and pH. Authors: Kayla Kindig / Eric Gibbs / David Seiferth / Philip C Biggin / Sudha Chakrapani /   Abstract: Glycine receptors (GlyRs) regulate motor control and pain processing in the central nervous system through inhibitory synaptic signaling. The subtype GlyRα3 expressed in nociceptive sensory neurons ...Glycine receptors (GlyRs) regulate motor control and pain processing in the central nervous system through inhibitory synaptic signaling. The subtype GlyRα3 expressed in nociceptive sensory neurons of the spinal dorsal horn is a key regulator of physiological pain perception. Disruption of spinal glycinergic inhibition is associated with chronic inflammatory pain states, making GlyRα3 an attractive target for pain treatment. GlyRα3 activity is modulated by numerous endogenous and exogenous ligands that consequently affect pain sensitization. To understand the mechanism of two such endogenous modulators, Zn and protons, we have used cryo-electron microscopy to determine structures of full-length human GlyRα3 in various functional states. Whereas acidic pH reduces peak glycine response, Zn displays biphasic modulation in a concentration-dependent manner. Our findings reveal the effector sites and also capture intermediate conformations in the gating cycle. Combined with molecular dynamics simulations and electrophysiology, this work provides important insights into GlyRα3 activation and regulation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44964.map.gz emd_44964.map.gz | 80.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44964-v30.xml emd-44964-v30.xml emd-44964.xml emd-44964.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44964_fsc.xml emd_44964_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_44964.png emd_44964.png | 97.4 KB | ||
| Filedesc metadata |  emd-44964.cif.gz emd-44964.cif.gz | 6.5 KB | ||
| Others |  emd_44964_half_map_1.map.gz emd_44964_half_map_1.map.gz emd_44964_half_map_2.map.gz emd_44964_half_map_2.map.gz | 81.1 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44964 http://ftp.pdbj.org/pub/emdb/structures/EMD-44964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44964 | HTTPS FTP |
-Related structure data
| Related structure data |  9bwcMC  9bu2C  9bu3C  9bvhC  9bvjC  9bwbC  9bweC  9bwgC  9bwjC  9bzpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44964.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44964.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_44964_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_44964_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pentameric Assembly of Human Glycine Receptor Subtype Alpha3
| Entire | Name: Pentameric Assembly of Human Glycine Receptor Subtype Alpha3 |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric Assembly of Human Glycine Receptor Subtype Alpha3
| Supramolecule | Name: Pentameric Assembly of Human Glycine Receptor Subtype Alpha3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Glycine receptor subunit alpha-3
| Macromolecule | Name: Glycine receptor subunit alpha-3 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.581066 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MAHVRHFRTL VSGFYFWEAA LLLSLVATKE TDSARSRSAP MSPSDFLDKL MGRTSGYDAR IRPNFKGPPV NVTCNIFINS FGSIAETTM DYRVNIFLRQ KWNDPRLAYS EYPDDSLDLD PSMLDSIWKP DLFFANEKGA NFHEVTTDNK LLRIFKNGNV L YSIRLTLT ...String: MAHVRHFRTL VSGFYFWEAA LLLSLVATKE TDSARSRSAP MSPSDFLDKL MGRTSGYDAR IRPNFKGPPV NVTCNIFINS FGSIAETTM DYRVNIFLRQ KWNDPRLAYS EYPDDSLDLD PSMLDSIWKP DLFFANEKGA NFHEVTTDNK LLRIFKNGNV L YSIRLTLT LSCPMDLKNF PMDVQTCIMQ LESFGYTMND LIFEWQDEAP VQVAEGLTLP QFLLKEEKDL RYCTKHYNTG KF TCIEVRF HLERQMGYYL IQMYIPSLLI VILSWVSFWI NMDAAPARVA LGITTVLTMT TQSSGSRASL PKVSYVKAID IWM AVCLLF VFSALLEYAA VNFVSRQHKE LLRFRRKRKN KTEAFALEKF YRFSDMDDEV RESRFSFTAY GMGPCLQAKD GMTP KGPNH PVQVMPKSPD EMRKVFIDRA KKIDTISRAC FPLAFLIFNI FYWVIYKILR HEDIHQQQDL VPRGSHHHHH HHH UniProtKB: Glycine receptor subunit alpha-3 |
-Macromolecule #3: 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 25 / Formula: PX4 |
|---|---|
| Molecular weight | Theoretical: 678.94 Da |
| Chemical component information |  ChemComp-PX4: |
-Macromolecule #4: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
| Macromolecule | Name: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate type: ligand / ID: 4 / Number of copies: 5 / Formula: PIO |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information |  ChemComp-PIO: |
-Macromolecule #5: GLYCINE
| Macromolecule | Name: GLYCINE / type: ligand / ID: 5 / Number of copies: 5 / Formula: GLY |
|---|---|
| Molecular weight | Theoretical: 75.067 Da |
| Chemical component information |  ChemComp-GLY: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 35 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)