+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4487 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of E. coli ribosome deposited by spraying | |||||||||
 Map data Map data | LocalRes filtered map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Kontziampasis D / Klebl DP / Iadanza MG / Scarff CA / Kopf F / Sobott F / Monteiro DCF / Trebbin M / Muench SP / White HD | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2019 Journal: IUCrJ / Year: 2019Title: A cryo-EM grid preparation device for time-resolved structural studies. Authors: Dimitrios Kontziampasis / David P Klebl / Matthew G Iadanza / Charlotte A Scarff / Florian Kopf / Frank Sobott / Diana C F Monteiro / Martin Trebbin / Stephen P Muench / Howard D White /     Abstract: Structural biology generally provides static snapshots of protein conformations that can provide information on the functional mechanisms of biological systems. Time-resolved structural biology ...Structural biology generally provides static snapshots of protein conformations that can provide information on the functional mechanisms of biological systems. Time-resolved structural biology provides a means to visualize, at near-atomic resolution, the dynamic conformational changes that macromolecules undergo as they function. X-ray free-electron-laser technology has provided a powerful tool to study enzyme mechanisms at atomic resolution, typically in the femtosecond to picosecond timeframe. Complementary to this, recent advances in the resolution obtainable by electron microscopy and the broad range of samples that can be studied make it ideally suited to time-resolved approaches in the microsecond to millisecond timeframe to study large loop and domain motions in biomolecules. Here we describe a cryo-EM grid preparation device that permits rapid mixing, voltage-assisted spraying and vitrification of samples. It is shown that the device produces grids of sufficient ice quality to enable data collection from single grids that results in a sub-4 Å reconstruction. Rapid mixing can be achieved by blot-and-spray or mix-and-spray approaches with a delay of ∼10 ms, providing greater temporal resolution than previously reported mix-and-spray approaches. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4487.map.gz emd_4487.map.gz | 61.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4487-v30.xml emd-4487-v30.xml emd-4487.xml emd-4487.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4487_fsc.xml emd_4487_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_4487.png emd_4487.png | 84.4 KB | ||
| Others |  emd_4487_additional_1.map.gz emd_4487_additional_1.map.gz emd_4487_additional_2.map.gz emd_4487_additional_2.map.gz emd_4487_half_map_1.map.gz emd_4487_half_map_1.map.gz emd_4487_half_map_2.map.gz emd_4487_half_map_2.map.gz | 22.9 MB 79.7 MB 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4487 http://ftp.pdbj.org/pub/emdb/structures/EMD-4487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4487 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4487.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4487.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocalRes filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
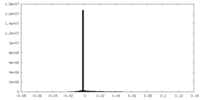
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map
| File | emd_4487_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
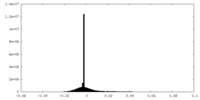
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_4487_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap 1
| File | emd_4487_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap 2
| File | emd_4487_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ribosome from E. coli
| Entire | Name: ribosome from E. coli |
|---|---|
| Components |
|
-Supramolecule #1: ribosome from E. coli
| Supramolecule | Name: ribosome from E. coli / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.9 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)