+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | WT Pseudomonas phage PP7 capped tube VLP | |||||||||
 Map data Map data | sharpened b-factor -47 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VLP / PP7 / coat / tube / VIRUS LIKE PARTICLE | |||||||||
| Biological species |  Pepevirus rubrum Pepevirus rubrum | |||||||||
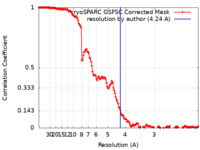
| Method | single particle reconstruction / cryo EM / Resolution: 4.24 Å | |||||||||
 Authors Authors | Kopylov M / Hernandez C / Bobe D / Keshavarz-Joud P / Finn MG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Small / Year: 2025 Journal: Small / Year: 2025Title: Pleomorphism in Wild-Type and Engineered PP7 Virus-Like Particles. Authors: Parisa Keshavarz-Joud / Matthew C Jenkins / Tahiti Dutta / Liangjun Zhao / Carolina Hernandez / Daija Bobe / Mohammadreza Paraan / M G Finn / Mykhailo Kopylov /  Abstract: Virus-like particles (VLPs) find applications across many different fields, aided by their stability, capacity for large-scale production, and presumed structural homogeneity. These attributes are a ...Virus-like particles (VLPs) find applications across many different fields, aided by their stability, capacity for large-scale production, and presumed structural homogeneity. These attributes are a result of their highly efficient self-assembly, which stems from the evolutionary pressures on the natural viruses from which they are derived. It is found that VLPs based on the Leviphage PP7 assemble in an unexpectedly wide range of morphologies. The relative abundance of these structures is sensitive to small changes in the coat protein sequence. These results raise the possibility that structural plasticity may be a general property of such self-assembling structures rather than an exception. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44737.map.gz emd_44737.map.gz | 266.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44737-v30.xml emd-44737-v30.xml emd-44737.xml emd-44737.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44737_fsc.xml emd_44737_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_44737.png emd_44737.png | 74.5 KB | ||
| Masks |  emd_44737_msk_1.map emd_44737_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44737.cif.gz emd-44737.cif.gz | 4.2 KB | ||
| Others |  emd_44737_additional_1.map.gz emd_44737_additional_1.map.gz emd_44737_half_map_1.map.gz emd_44737_half_map_1.map.gz emd_44737_half_map_2.map.gz emd_44737_half_map_2.map.gz | 140.9 MB 262 MB 262 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44737 http://ftp.pdbj.org/pub/emdb/structures/EMD-44737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44737 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44737.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44737.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened b-factor -47 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
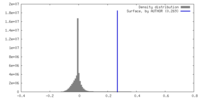
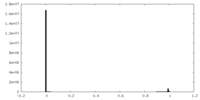
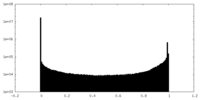
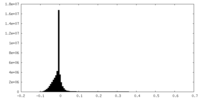
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44737_msk_1.map emd_44737_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
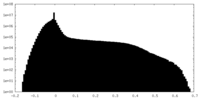
| Density Histograms |
-Additional map: unsharpened
| File | emd_44737_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
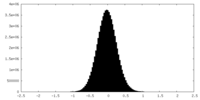
| Density Histograms |
-Half map: #1
| File | emd_44737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
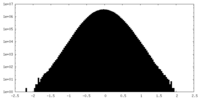
| Density Histograms |
-Half map: #2
| File | emd_44737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
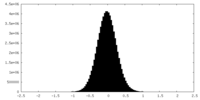
| Density Histograms |
- Sample components
Sample components
-Entire : C5 capped tube self-assembly of wild type PP7 bacteriophage prote...
| Entire | Name: C5 capped tube self-assembly of wild type PP7 bacteriophage protein expressedin E.coli |
|---|---|
| Components |
|
-Supramolecule #1: C5 capped tube self-assembly of wild type PP7 bacteriophage prote...
| Supramolecule | Name: C5 capped tube self-assembly of wild type PP7 bacteriophage protein expressedin E.coli type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Pepevirus rubrum Pepevirus rubrum |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 0.1 M / Component - Name: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER / Details: Solarus, 15W |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)