[English] 日本語
 Yorodumi
Yorodumi- EMDB-44134: type 2 KD-mxyl filament of miniature tau macrocycle derived from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold | |||||||||
 Map data Map data | type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tauopathies / neurodegenerative disorders / seed-competent miniature tau macrocycle / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / negative regulation of tubulin deacetylation ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / negative regulation of tubulin deacetylation / generation of neurons / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / regulation of chromosome organization / central nervous system neuron development / intracellular distribution of mitochondria / minor groove of adenine-thymine-rich DNA binding / lipoprotein particle binding / microtubule polymerization / negative regulation of mitochondrial membrane potential / regulation of microtubule polymerization / dynactin binding / apolipoprotein binding / main axon / protein polymerization / glial cell projection / axolemma / negative regulation of mitochondrial fission / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / neurofibrillary tangle assembly / positive regulation of axon extension / Activation of AMPK downstream of NMDARs / regulation of cellular response to heat / positive regulation of superoxide anion generation / synapse assembly / supramolecular fiber organization / regulation of long-term synaptic depression / positive regulation of protein localization / cellular response to brain-derived neurotrophic factor stimulus / regulation of calcium-mediated signaling / cytoplasmic microtubule organization / positive regulation of microtubule polymerization / somatodendritic compartment / axon cytoplasm / astrocyte activation / stress granule assembly / phosphatidylinositol binding / nuclear periphery / regulation of microtubule cytoskeleton organization / protein phosphatase 2A binding / cellular response to reactive oxygen species / Hsp90 protein binding / microglial cell activation / synapse organization / PKR-mediated signaling / cellular response to nerve growth factor stimulus / protein homooligomerization / regulation of synaptic plasticity / SH3 domain binding / response to lead ion / microtubule cytoskeleton organization / memory / cytoplasmic ribonucleoprotein granule / neuron projection development / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / actin binding / cellular response to heat / growth cone / microtubule cytoskeleton / cell body / double-stranded DNA binding / protein-macromolecule adaptor activity / microtubule binding / dendritic spine / sequence-specific DNA binding / amyloid fibril formation / microtubule / learning or memory / neuron projection / regulation of autophagy / membrane raft / axon / negative regulation of gene expression / neuronal cell body / dendrite / DNA damage response / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Xu X / Angera JI / Rajewski HB / Jiang W / Del Valle RJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure-based design of seed-competent proteomimetic macrocycles derived from 4R tauopathic folds Authors: Xu X / Angera JI / Rajewski HB / Jiang W / Del Valle RJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44134.map.gz emd_44134.map.gz | 116.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44134-v30.xml emd-44134-v30.xml emd-44134.xml emd-44134.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44134_fsc.xml emd_44134_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_44134.png emd_44134.png | 135.3 KB | ||
| Filedesc metadata |  emd-44134.cif.gz emd-44134.cif.gz | 5.1 KB | ||
| Others |  emd_44134_half_map_1.map.gz emd_44134_half_map_1.map.gz emd_44134_half_map_2.map.gz emd_44134_half_map_2.map.gz | 98.4 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44134 http://ftp.pdbj.org/pub/emdb/structures/EMD-44134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44134 | HTTPS FTP |
-Validation report
| Summary document |  emd_44134_validation.pdf.gz emd_44134_validation.pdf.gz | 942.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44134_full_validation.pdf.gz emd_44134_full_validation.pdf.gz | 942.2 KB | Display | |
| Data in XML |  emd_44134_validation.xml.gz emd_44134_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_44134_validation.cif.gz emd_44134_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44134 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44134 | HTTPS FTP |
-Related structure data
| Related structure data |  9b3cMC  9b3aC  9dmeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44134.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44134.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map A
| File | emd_44134_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_44134_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : type 2 KD-mxyl filament of miniature tau macrocycle derived from ...
| Entire | Name: type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold |
|---|---|
| Components |
|
-Supramolecule #1: type 2 KD-mxyl filament of miniature tau macrocycle derived from ...
| Supramolecule | Name: type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Microtubule-associated protein tau
| Macromolecule | Name: Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.24264 KDa |
| Sequence | String: (ACE)CDNIKHVPG GGSVQIVYKP VC UniProtKB: Microtubule-associated protein tau |
-Macromolecule #2: GLY-SER-VAL-GLN-ILE-VAL-TYR
| Macromolecule | Name: GLY-SER-VAL-GLN-ILE-VAL-TYR / type: protein_or_peptide / ID: 2 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 764.867 Da |
| Sequence | String: GSVQIVY |
-Macromolecule #3: AMINO GROUP
| Macromolecule | Name: AMINO GROUP / type: ligand / ID: 3 / Number of copies: 15 / Formula: NH2 |
|---|---|
| Molecular weight | Theoretical: 16.023 Da |
| Chemical component information |  ChemComp-NH2: |
-Macromolecule #4: 1,3-dimethylbenzene
| Macromolecule | Name: 1,3-dimethylbenzene / type: ligand / ID: 4 / Number of copies: 15 / Formula: 8VH |
|---|---|
| Molecular weight | Theoretical: 106.165 Da |
| Chemical component information | 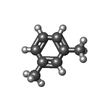 ChemComp-8VH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.495 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-9b3c: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































