+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bovine fetal muscle nAChR bound to ACh | |||||||||
 Map data Map data | sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bovine muscle nicotinic acetylcholine receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHighly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / Highly calcium permeable nicotinic acetylcholine receptors / postsynaptic membrane organization / skeletal muscle tissue growth / musculoskeletal movement / acetylcholine receptor activity / excitatory extracellular ligand-gated monoatomic ion channel activity / acetylcholine-gated channel complex / behavioral response to nicotine ...Highly sodium permeable postsynaptic acetylcholine nicotinic receptors / Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / Highly calcium permeable nicotinic acetylcholine receptors / postsynaptic membrane organization / skeletal muscle tissue growth / musculoskeletal movement / acetylcholine receptor activity / excitatory extracellular ligand-gated monoatomic ion channel activity / acetylcholine-gated channel complex / behavioral response to nicotine / neuromuscular synaptic transmission / acetylcholine-gated monoatomic cation-selective channel activity / muscle cell development / acetylcholine binding / synaptic transmission, cholinergic / nervous system process / acetylcholine receptor signaling pathway / postsynaptic specialization membrane / membrane depolarization / monoatomic cation transport / muscle contraction / response to nicotine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / neuromuscular junction / transmembrane signaling receptor activity / channel activity / monoatomic ion transmembrane transport / chemical synaptic transmission / postsynaptic membrane / neuron projection / synapse / signal transduction / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
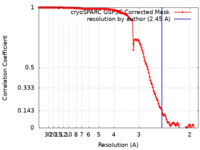
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | |||||||||
 Authors Authors | Li H / Hibbs RE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural switch in acetylcholine receptors in developing muscle. Authors: Huanhuan Li / Jinfeng Teng / Ryan E Hibbs /  Abstract: During development, motor neurons originating in the brainstem and spinal cord form elaborate synapses with skeletal muscle fibres. These neurons release acetylcholine (ACh), which binds to nicotinic ...During development, motor neurons originating in the brainstem and spinal cord form elaborate synapses with skeletal muscle fibres. These neurons release acetylcholine (ACh), which binds to nicotinic ACh receptors (AChRs) on the muscle, initiating contraction. Two types of AChR are present in developing muscle cells, and their differential expression serves as a hallmark of neuromuscular synapse maturation. The structural principles underlying the switch from fetal to adult muscle receptors are unknown. Here, we present high-resolution structures of both fetal and adult muscle nicotinic AChRs, isolated from bovine skeletal muscle in developmental transition. These structures, obtained in the absence and presence of ACh, provide a structural context for understanding how fetal versus adult receptor isoforms are tuned for synapse development versus the all-or-none signalling required for high-fidelity skeletal muscle contraction. We find that ACh affinity differences are driven by binding site access, channel conductance is tuned by widespread surface electrostatics and open duration changes result from intrasubunit interactions and structural flexibility. The structures further reveal pathogenic mechanisms underlying congenital myasthenic syndromes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43923.map.gz emd_43923.map.gz | 226.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43923-v30.xml emd-43923-v30.xml emd-43923.xml emd-43923.xml | 28.1 KB 28.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43923_fsc.xml emd_43923_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_43923.png emd_43923.png | 42.6 KB | ||
| Filedesc metadata |  emd-43923.cif.gz emd-43923.cif.gz | 8 KB | ||
| Others |  emd_43923_additional_1.map.gz emd_43923_additional_1.map.gz emd_43923_additional_2.map.gz emd_43923_additional_2.map.gz emd_43923_half_map_1.map.gz emd_43923_half_map_1.map.gz emd_43923_half_map_2.map.gz emd_43923_half_map_2.map.gz | 206.6 MB 118.7 MB 222.8 MB 222.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43923 http://ftp.pdbj.org/pub/emdb/structures/EMD-43923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43923 | HTTPS FTP |
-Related structure data
| Related structure data |  9avuMC  9avvC  9awjC  9awkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43923.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43923.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharp | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
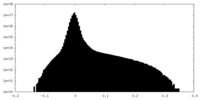
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: emhancer
| File | emd_43923_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | emhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
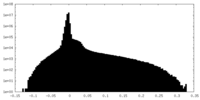
| Density Histograms |
-Additional map: unsharp
| File | emd_43923_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharp | ||||||||||||
| Projections & Slices |
| ||||||||||||
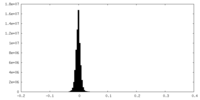
| Density Histograms |
-Half map: fetal desen halfmapA
| File | emd_43923_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | fetal_desen_halfmapA | ||||||||||||
| Projections & Slices |
| ||||||||||||
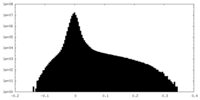
| Density Histograms |
-Half map: fetal desen halfmapB
| File | emd_43923_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | fetal_desen_halfmapB | ||||||||||||
| Projections & Slices |
| ||||||||||||
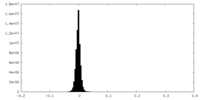
| Density Histograms |
- Sample components
Sample components
-Entire : Bovine muscle nicotinic acetylcholine receptor
| Entire | Name: Bovine muscle nicotinic acetylcholine receptor |
|---|---|
| Components |
|
-Supramolecule #1: Bovine muscle nicotinic acetylcholine receptor
| Supramolecule | Name: Bovine muscle nicotinic acetylcholine receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Acetylcholine receptor subunit alpha
| Macromolecule | Name: Acetylcholine receptor subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.949828 KDa |
| Sequence | String: SEHETRLVAK LFEDYNSVVR PVEDHRQAVE VTVGLQLIQL INVDEVNQIV TTNVRLKQQW VDYNLKWNPD DYGGVKKIHI PSEKIWRPD LVLYNNADGD FAIVKFTKVL LDYTGHITWT PPAIFKSYCE IIVTHFPFDE QNCSMKLGTW TYDGSVVVIN P ESDQPDLS ...String: SEHETRLVAK LFEDYNSVVR PVEDHRQAVE VTVGLQLIQL INVDEVNQIV TTNVRLKQQW VDYNLKWNPD DYGGVKKIHI PSEKIWRPD LVLYNNADGD FAIVKFTKVL LDYTGHITWT PPAIFKSYCE IIVTHFPFDE QNCSMKLGTW TYDGSVVVIN P ESDQPDLS NFMESGEWVI KESRGWKHWV FYACCPSTPY LDITYHFVMQ RLPLYFIVNV IIPCLLFSFL TGLVFYLPTD SG EKMTLSI SVLLSLTVFL LVIVELIPST SSAVPLIGKY MLFTMVFVIA SIIITVIVIN THHRSPSTHV MPEWVRKVFI DTI PNIMFF STMKRPSREK QDKKIFTEDI DISDISGKPG PPPMGFHSPL IKHPEVKSAI EGIKYIAETM KSDQESNNAA EEWK YVAMV MDHILLAVFM LVCIIGTLAV FAGRLIELNQ QG UniProtKB: Acetylcholine receptor subunit alpha |
-Macromolecule #2: Acetylcholine receptor subunit beta
| Macromolecule | Name: Acetylcholine receptor subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.111832 KDa |
| Sequence | String: SEAEGRLREK LFSGYDSTVR PAREVGDRVW VSIGLTLAQL ISLNEKDEEM STKVYLDLEW TDYRLSWDPE EHEGIDSLRI SAESVWLPD VVLLNNNDGN FDVALDINVV VSSDGSMRWQ PPGIYRSSCS IQVTYFPFDW QNCTMVFSSY SYDSSEVSLQ T GLSPEGQE ...String: SEAEGRLREK LFSGYDSTVR PAREVGDRVW VSIGLTLAQL ISLNEKDEEM STKVYLDLEW TDYRLSWDPE EHEGIDSLRI SAESVWLPD VVLLNNNDGN FDVALDINVV VSSDGSMRWQ PPGIYRSSCS IQVTYFPFDW QNCTMVFSSY SYDSSEVSLQ T GLSPEGQE RQEVYIHEGT FIENGQWEII HKPSRLIQPS VDPRGGGEGR REEVTFYLII RRKPLFYLVN VIAPCILITL LA IFVFYLP PDAGEKMGLS IFALLTLTVF LLLLADKVPE TSLSVPIIIK YLMFTMVLVT FSVILSVVVL NLHHRSPHTH QMP LWVRQI FIHKLPLYLG LKRPKPERDQ MQEPPSIAPR DSPGSGWGRG TDEYFIRKPP NDFLFPKPNR FQPELSAPDL RRFI DGPNR AVGLPPELRE VVSSISYIAR QLQEQEDHDV LKEDWQFVAM VVDRLFLWTF IIFTSVGTLV IFLDATYHLP PADPF P UniProtKB: Acetylcholine receptor subunit beta |
-Macromolecule #3: Acetylcholine receptor subunit delta
| Macromolecule | Name: Acetylcholine receptor subunit delta / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.543926 KDa |
| Sequence | String: LNEEERLIRH LFEEKAYNKE LRPAAHKESV EISLALTLSN LISLKEVEET LTTNVWIEQG WTDSRLQWDA EDFGNISVLR LPADMVWLP EIVLENNNDG SFQISYSCNV LIYPSGSVYW LPPAIFRSSC PISVTYFPFD WQNCSLKFSS LKYTTKEITL S LKQAEEDG ...String: LNEEERLIRH LFEEKAYNKE LRPAAHKESV EISLALTLSN LISLKEVEET LTTNVWIEQG WTDSRLQWDA EDFGNISVLR LPADMVWLP EIVLENNNDG SFQISYSCNV LIYPSGSVYW LPPAIFRSSC PISVTYFPFD WQNCSLKFSS LKYTTKEITL S LKQAEEDG RSYPVEWIII DPEGFTENGE WEIVHRPARV NVDPSVPLDS PNRQDVTFYL IIRRKPLFYV INILVPCVLI SF MINLVFY LPADCGEKTS MAISVLLAQS VFLLLISKRL PATSMAIPLI GKFLLFGMVL VTMVVVICVI VLNIHFRTPS THV LSEPVK KLFLETLPEI LHMSRPAEDG PSPGTLIRRS SSLGYISKAE EYFSLKSRSD LMFEKQSERH GLARRLTTAR RPPA GSEQA QQELFSELKP AVDGANFIVN HMKDQNNYNE EKDCWNRVAR TVDRLCLFVV TPIMVVGTAW IFLQGAYNQP PPQPF PGDP FSYLEKDKRF I UniProtKB: Acetylcholine receptor subunit delta |
-Macromolecule #4: Acetylcholine receptor subunit gamma
| Macromolecule | Name: Acetylcholine receptor subunit gamma / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.99468 KDa |
| Sequence | String: RNQEERLLGD LMQGYNPHLR PAEHDSDVVN VSLKLTLTNL ISLNEREEAL TTNVWIEMQW CDYRLRWDPR DYGGLWVLRV PSTMVWRPD IVLENNVDGV FEVALYCNVL VSPDGCVYWL PPAIFRSSCP VSVTFFPFDW QNCSLIFQSQ TYSTNEINLQ L SQEDGQTI ...String: RNQEERLLGD LMQGYNPHLR PAEHDSDVVN VSLKLTLTNL ISLNEREEAL TTNVWIEMQW CDYRLRWDPR DYGGLWVLRV PSTMVWRPD IVLENNVDGV FEVALYCNVL VSPDGCVYWL PPAIFRSSCP VSVTFFPFDW QNCSLIFQSQ TYSTNEINLQ L SQEDGQTI EWIFIDPEAF TENGEWAIRH RPAKMLLDEA APAEEAGHQK VVFYLLIQRK PLFYVINIIA PCVLISSVAI LI YFLPAKA GGQKCTVAIN VLLAQTVFLF LVAKKVPETS QAVPLISKYL TFLLVVTILI VVNAVVVLNV SLRSPHTHSM ARG VRKVFL RLLPQLLRMH VRPLAPVAVQ DAHPRLQNGS SSGWPITAGE EVALCLPRSE LLFRQRQRNG LVRAALEKLE KGPE SGQSP EWCGSLKQAA PAIQACVEAC NLIARARHQQ THFDSGNKEW FLVGRVLDRV CFLAMLSLFV CGTAGIFLMA HYNRV PALP FPGDPRSYLP SSD UniProtKB: Acetylcholine receptor subunit gamma |
-Macromolecule #10: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 10 / Number of copies: 2 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #11: ACETYLCHOLINE
| Macromolecule | Name: ACETYLCHOLINE / type: ligand / ID: 11 / Number of copies: 2 / Formula: ACH |
|---|---|
| Molecular weight | Theoretical: 146.207 Da |
| Chemical component information | 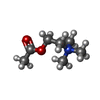 ChemComp-ACH: |
-Macromolecule #12: water
| Macromolecule | Name: water / type: ligand / ID: 12 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: experimental model / Details: cryosparc ab initial model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-9avu: |
 Movie
Movie Controller
Controller



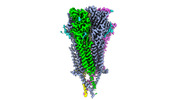





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)