+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human OGG1 bound at the nucleosomal super helical location -5 | |||||||||
 Map data Map data | Human OGG1 binds nucleosome at SHL -5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleosome core particle containing 8-oxoG DNA damage / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||
 Authors Authors | You Q / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations. Authors: Qinglong You / Xiang Feng / Yi Cai / Stephen B Baylin / Huilin Li /  Abstract: The human glycosylase OGG1 extrudes and excises the oxidized DNA base 8-oxoguanine (8-oxoG) to initiate base excision repair and plays important roles in many pathological conditions such as cancer, ...The human glycosylase OGG1 extrudes and excises the oxidized DNA base 8-oxoguanine (8-oxoG) to initiate base excision repair and plays important roles in many pathological conditions such as cancer, inflammation, and neurodegenerative diseases. Previous structural studies have used a truncated protein and short linear DNA, so it has been unclear how full-length OGG1 operates on longer DNA or on nucleosomes. Here we report cryo-EM structures of human OGG1 bound to a 35-bp long DNA containing an 8-oxoG within an unmethylated Cp-8-oxoG dinucleotide as well as to a nucleosome with an 8-oxoG at super-helical location (SHL)-5. The 8-oxoG in the linear DNA is flipped out by OGG1, consistent with previous crystallographic findings with a 15-bp DNA. OGG1 preferentially binds near dsDNA ends at the nucleosomal entry/exit sites. Such preference may underlie the enzyme's function in DNA double-strand break repair. Unexpectedly, we find that OGG1 bends the nucleosomal entry DNA, flips an undamaged guanine, and binds to internal nucleosomal DNA sites such as SHL-5 and SHL+6. We suggest that the DNA base search mechanism by OGG1 may be chromatin context-dependent and that OGG1 may partner with chromatin remodelers to excise 8-oxoG at the nucleosomal internal sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43610.map.gz emd_43610.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43610-v30.xml emd-43610-v30.xml emd-43610.xml emd-43610.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43610.png emd_43610.png | 56.4 KB | ||
| Filedesc metadata |  emd-43610.cif.gz emd-43610.cif.gz | 4.8 KB | ||
| Others |  emd_43610_half_map_1.map.gz emd_43610_half_map_1.map.gz emd_43610_half_map_2.map.gz emd_43610_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43610 http://ftp.pdbj.org/pub/emdb/structures/EMD-43610 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43610 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43610 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43610.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43610.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human OGG1 binds nucleosome at SHL -5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.656 Å | ||||||||||||||||||||||||||||||||||||
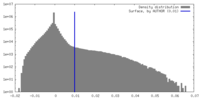
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human OGG1 binds nucleosome at SHL -5 half map 1
| File | emd_43610_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human OGG1 binds nucleosome at SHL -5 half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
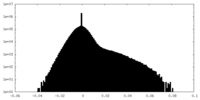
| Density Histograms |
-Half map: Human OGG1 binds nucleosome at SHL -5 half map 2
| File | emd_43610_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human OGG1 binds nucleosome at SHL -5 half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG
| Entire | Name: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG
| Supramolecule | Name: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: OGG1
| Macromolecule | Name: OGG1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHDY KDHDGDYKDH DIDYKDDDDK ENLYFQGGGG GSDPARALLP RRMGHRTLAS TPALWASIPC PRSELRLDLV LPSGQSFRWR EQSPAHWSGV LADQVWTLTQ TEEQLHCTVY RGDKSQASRP TPDELEAVRK YFQLDVTLAQ LYHHWGSVDS HFQEVAQKFQ ...String: MGHHHHHHDY KDHDGDYKDH DIDYKDDDDK ENLYFQGGGG GSDPARALLP RRMGHRTLAS TPALWASIPC PRSELRLDLV LPSGQSFRWR EQSPAHWSGV LADQVWTLTQ TEEQLHCTVY RGDKSQASRP TPDELEAVRK YFQLDVTLAQ LYHHWGSVDS HFQEVAQKFQ GVRLLRQDPI ECLFSFICSS NNNIARITGM VERLCQAFGP RLIQLDDVTY HGFPSLQALA GPEVEAHLRK LGLGYRARYV SASARAILEE QGGLAWLQQL RESSYEEAHK ALCILPGVGT QVADCICLMA LDKPQAVPVD VHMWHIAQRD YSWHPTTSQA KGPSPQTNKE LGNFFRSLWG PYAGWAQAVL FSADLRQSRH AQEPPAKRRK GSKGPEG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 51710 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)