+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleosome core particle containing an 8-oxoG damage site | |||||||||
 Map data Map data | Nucleosome core particle containing 8-oxoG DNA damage | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleosome core particle containing 8-oxoG DNA damage / STRUCTURAL PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | You Q / Li H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations. Authors: Qinglong You / Xiang Feng / Yi Cai / Stephen B Baylin / Huilin Li /  Abstract: The human glycosylase OGG1 extrudes and excises the oxidized DNA base 8-oxoguanine (8-oxoG) to initiate base excision repair and plays important roles in many pathological conditions such as cancer, ...The human glycosylase OGG1 extrudes and excises the oxidized DNA base 8-oxoguanine (8-oxoG) to initiate base excision repair and plays important roles in many pathological conditions such as cancer, inflammation, and neurodegenerative diseases. Previous structural studies have used a truncated protein and short linear DNA, so it has been unclear how full-length OGG1 operates on longer DNA or on nucleosomes. Here we report cryo-EM structures of human OGG1 bound to a 35-bp long DNA containing an 8-oxoG within an unmethylated Cp-8-oxoG dinucleotide as well as to a nucleosome with an 8-oxoG at super-helical location (SHL)-5. The 8-oxoG in the linear DNA is flipped out by OGG1, consistent with previous crystallographic findings with a 15-bp DNA. OGG1 preferentially binds near dsDNA ends at the nucleosomal entry/exit sites. Such preference may underlie the enzyme's function in DNA double-strand break repair. Unexpectedly, we find that OGG1 bends the nucleosomal entry DNA, flips an undamaged guanine, and binds to internal nucleosomal DNA sites such as SHL-5 and SHL+6. We suggest that the DNA base search mechanism by OGG1 may be chromatin context-dependent and that OGG1 may partner with chromatin remodelers to excise 8-oxoG at the nucleosomal internal sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43608.map.gz emd_43608.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43608-v30.xml emd-43608-v30.xml emd-43608.xml emd-43608.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43608.png emd_43608.png | 95.7 KB | ||
| Filedesc metadata |  emd-43608.cif.gz emd-43608.cif.gz | 6.6 KB | ||
| Others |  emd_43608_half_map_1.map.gz emd_43608_half_map_1.map.gz emd_43608_half_map_2.map.gz emd_43608_half_map_2.map.gz | 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43608 http://ftp.pdbj.org/pub/emdb/structures/EMD-43608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43608 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43608 | HTTPS FTP |
-Validation report
| Summary document |  emd_43608_validation.pdf.gz emd_43608_validation.pdf.gz | 807 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43608_full_validation.pdf.gz emd_43608_full_validation.pdf.gz | 806.5 KB | Display | |
| Data in XML |  emd_43608_validation.xml.gz emd_43608_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_43608_validation.cif.gz emd_43608_validation.cif.gz | 12.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43608 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43608 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43608 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43608 | HTTPS FTP |
-Related structure data
| Related structure data |  8vx5MC  8vx4C  8vx6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43608.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43608.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleosome core particle containing 8-oxoG DNA damage | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
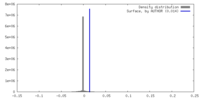
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Nucleosome core particle containing 8-oxoG DNA damage half map 1
| File | emd_43608_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleosome core particle containing 8-oxoG DNA damage half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
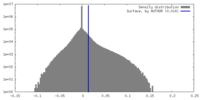
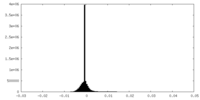
| Density Histograms |
-Half map: Nucleosome core particle containing 8-oxoG DNA damage half map 2
| File | emd_43608_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nucleosome core particle containing 8-oxoG DNA damage half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
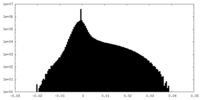
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG
| Entire | Name: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG
| Supramolecule | Name: Complex of Human OGG1 with a 35 bp DNA duplex containing 8-oxoG type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone H3.2
| Macromolecule | Name: Histone H3.2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.435126 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEASEA YLVALFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.2 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.307514 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGGSSGLVP RGSLEHHHHH H UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 18.086951 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKETAAAKFE RQHMDSPDLH HHHHHGTLVP RGSMGMSGRG KQGGKTRAKA KTRSSRAGLQ FPVGRVHRLL RKGNYAERVG AGAPVYLAA VLEYLTAEIL ELAGNAARDN KKTRIIPRHL QLAVRNDEEL NKLLGRVTIA QGGVLPNIQS VLLPKKTESS K SAKSK UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.655948 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKSAPAPKK GSKKAVTKTQ KKDGKKRRKT RKESYAIYVY KVLKQVHPDT GISSKAMSIM NSFVNDVFER IAGEASRLAH YNKRSTITS REIQTAVRLL LPGELAKHAV SEGTKAVTKY TSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #5: DNA (167-MER)
| Macromolecule | Name: DNA (167-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 51.785953 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DC)(DC)(DG)(DC)(DC) (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA) (DC)(DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG) (DA)(DG)(DA)(DC)(DT)(DA) ...String: (DA)(DT)(DC)(DG)(DG)(DC)(DC)(DG)(DC)(DC) (DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA) (DC)(DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG) (DG)(DA)(DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT) (DT)(DG)(DG)(DC)(DG)(DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG) (DA)(DC)(DA)(DG)(DC)(DG) (DC)(DG)(DT)(DA)(DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA) (DA)(DG)(DC)(DG)(DG) (DT)(DG)(DC)(DT)(DA)(DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA) (DC)(DG)(DA)(DC) (DC)(DA)(DA)(DT)(DT)(DG)(DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG) (DG)(DC)(DA) (DC)(DC)(DG)(DG)(DG)(DA)(DT)(DT)(DC)(DT) (DC)(DC)(DA)(DG)(DG)(DG)(DC) (DG)(DG) (DC)(DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (167-MER)
| Macromolecule | Name: DNA (167-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 51.343652 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DC)(DC)(DG)(DC)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG) (DA)(DG)(DG)(DC)(DC)(8OG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DC)(DC)(DG)(DC)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG) (DA)(DG)(DG)(DC)(DC)(8OG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT) (DA)(DG)(DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT) (DT)(DA)(DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA) (DC)(DG)(DC)(DG)(DC)(DT)(DG) (DT)(DC)(DC)(DC)(DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DT) (DA)(DA)(DC)(DC)(DG)(DC) (DC)(DA)(DA)(DG)(DG)(DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC) (DC)(DC)(DT)(DA)(DG) (DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT) (DC)(DA)(DG)(DA) (DT)(DA)(DT)(DA)(DT)(DA)(DC)(DA)(DT)(DC) (DC)(DT)(DG)(DT)(DG)(DG) (DC)(DG)(DG) (DC)(DC)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8vx5: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)