+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AaegOR10 structure bound to o-cresol | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mosquito olfactory receptor OR10 / MEMBRANE PROTEIN | |||||||||
| Function / homology | Olfactory receptor, insect / 7tm Odorant receptor / olfactory receptor activity / odorant binding / signal transduction / identical protein binding / plasma membrane / Odorant receptor / Odorant receptor Function and homology information Function and homology information | |||||||||
| Biological species |   Apocrypta bakeri (insect) Apocrypta bakeri (insect) | |||||||||
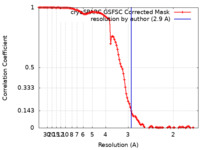
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Zhao J / del Marmol J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Structural basis of odor sensing by insect heteromeric odorant receptors. Authors: Jiawei Zhao / Andy Q Chen / Jaewook Ryu / Josefina Del Mármol /  Abstract: Most insects, including human-targeting mosquitoes, detect odors through odorant-activated ion channel complexes consisting of a divergent odorant-binding subunit (OR) and a conserved co-receptor ...Most insects, including human-targeting mosquitoes, detect odors through odorant-activated ion channel complexes consisting of a divergent odorant-binding subunit (OR) and a conserved co-receptor subunit (Orco). As a basis for understanding how odorants activate these heteromeric receptors, we report here cryo-electron microscopy structures of two different heteromeric odorant receptor complexes containing ORs from disease-vector mosquitos or . These structures reveal an unexpected stoichiometry of one OR to three Orco subunits. Comparison of structures in odorant-bound and unbound states indicates that odorant binding to the sole OR subunit is sufficient to open the channel pore, suggesting a mechanism of OR activation and a conceptual framework for understanding evolution of insect odorant receptor sensitivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42850.map.gz emd_42850.map.gz | 129.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42850-v30.xml emd-42850-v30.xml emd-42850.xml emd-42850.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42850_fsc.xml emd_42850_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_42850.png emd_42850.png | 170.4 KB | ||
| Filedesc metadata |  emd-42850.cif.gz emd-42850.cif.gz | 5.9 KB | ||
| Others |  emd_42850_half_map_1.map.gz emd_42850_half_map_1.map.gz emd_42850_half_map_2.map.gz emd_42850_half_map_2.map.gz | 127.2 MB 127.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42850 http://ftp.pdbj.org/pub/emdb/structures/EMD-42850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42850 | HTTPS FTP |
-Related structure data
| Related structure data |  8v02MC  8v00C  8v3cC  8v3dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42850.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42850.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
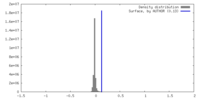
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42850_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
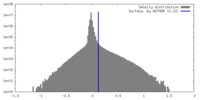
| Density Histograms |
-Half map: #1
| File | emd_42850_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
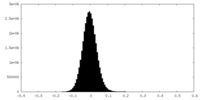
| Density Histograms |
- Sample components
Sample components
-Entire : AaegOR10 heterotetramer bound to o-cresol
| Entire | Name: AaegOR10 heterotetramer bound to o-cresol |
|---|---|
| Components |
|
-Supramolecule #1: AaegOR10 heterotetramer bound to o-cresol
| Supramolecule | Name: AaegOR10 heterotetramer bound to o-cresol / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Aedes aegypti OR10
| Supramolecule | Name: Aedes aegypti OR10 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Apocrypta bakeri Orco
| Supramolecule | Name: Apocrypta bakeri Orco / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Apocrypta bakeri (insect) Apocrypta bakeri (insect) |
-Macromolecule #1: Odorant receptor OR10
| Macromolecule | Name: Odorant receptor OR10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.491168 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASILDCPIV SVNARVWRFW SFVLKHDAMR YISIIPVTVM TFFMFTDLCR SWGNIQELII KAYFAVLYFN AVLRTLILVK DRKLYENFM QGISNVYFEI SHIDDHKIQS LLKSYTVRAR MLSISNLALG AIISTCFVVY PIFTGERGLP YGMFIPGLDS F RSPHYEII ...String: MASILDCPIV SVNARVWRFW SFVLKHDAMR YISIIPVTVM TFFMFTDLCR SWGNIQELII KAYFAVLYFN AVLRTLILVK DRKLYENFM QGISNVYFEI SHIDDHKIQS LLKSYTVRAR MLSISNLALG AIISTCFVVY PIFTGERGLP YGMFIPGLDS F RSPHYEII YIVQVVLTFP GCCMYIPFTS FFASTTLFGL VQIKTLQRQL QTFKDNINSQ DKEKVKAKVV KLIEDHKRII TY VSELNSL VTYICFVEFL SFGMMLCALL FLLNVIENHA QIVIVAAYIF MIISQIFAFY WHANEVREES MNLAEAAYSG PWV ELDNSI KKKLLLIILR AQQPLEITVG NVYPMTLEMF QSLLNASYSY FTLLRRVYN UniProtKB: Odorant receptor |
-Macromolecule #2: Odorant receptor Orco
| Macromolecule | Name: Odorant receptor Orco / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Apocrypta bakeri (insect) Apocrypta bakeri (insect) |
| Molecular weight | Theoretical: 53.204953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKFKHQGLVA DLLPNIRVMQ GVGHFMFNYY SEGKKFPHRI YCIVTLLLLL LQYGMMAVNL MMESDDVDDL TANTITMLFF LHPIVKMIY FPVRSKIFYK TLAIWNNPNS HPLFAESNAR FHALAITKMR RLLFCVAGAT IFSVISWTGI TFIEDSVKRI T DPETNETT ...String: MKFKHQGLVA DLLPNIRVMQ GVGHFMFNYY SEGKKFPHRI YCIVTLLLLL LQYGMMAVNL MMESDDVDDL TANTITMLFF LHPIVKMIY FPVRSKIFYK TLAIWNNPNS HPLFAESNAR FHALAITKMR RLLFCVAGAT IFSVISWTGI TFIEDSVKRI T DPETNETT IIPIPRLMIR TFYPFNAMSG AGHVFALIYQ FYYLVISMAV SNSLDVLFCS WLLFACEQLQ HLKAIMKPLM EL SATLDTV VPNSGELFKA GSADHLRESQ GVQPSGNGDN VLDVDLRGIY SNRQDFTATF RPTAGTTFNG GVGPNGLTKK QEM LVRSAI KYWVERHKHV VRLVTAVGDA YGVALLLHML TTTITLTLLA YQATKVNGVN VYAATVIGYL LYTLGQVFLF CIFG NRLIE ESSSVMEAAY SCHWYDGSEE AKTFVQIVCQ QCQKAMSISG AKFFTVSLDL FASVLGAVVT YFMVLVQLK UniProtKB: Odorant receptor |
-Macromolecule #3: o-cresol
| Macromolecule | Name: o-cresol / type: ligand / ID: 3 / Number of copies: 1 / Formula: JZ0 |
|---|---|
| Molecular weight | Theoretical: 108.138 Da |
| Chemical component information | 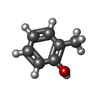 ChemComp-JZ0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Average electron dose: 55.2 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 58.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)