[English] 日本語
 Yorodumi
Yorodumi- EMDB-42779: Cryo-EM structure of a bacterial nitrilase filament with a covale... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a bacterial nitrilase filament with a covalent adduct derived from benzonitrile hydrolysis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Aromatic-nitrilase / helical-filament / benzaldehyde covalent-adduct intermediate / truncated mutant / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria) Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria) | |||||||||
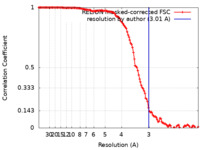
| Method | helical reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Aguirre-Sampieri S / Casanal A / Emsley P / Garza-Ramos G | |||||||||
| Funding support |  Mexico, 1 items Mexico, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2024 Journal: J Struct Biol / Year: 2024Title: Cryo-EM structure of bacterial nitrilase reveals insight into oligomerization, substrate recognition, and catalysis. Authors: Sergio Aguirre-Sampieri / Ana Casañal / Paul Emsley / Georgina Garza-Ramos /    Abstract: Many enzymes can self-assemble into higher-order structures with helical symmetry. A particularly noteworthy example is that of nitrilases, enzymes in which oligomerization of dimers into spiral homo- ...Many enzymes can self-assemble into higher-order structures with helical symmetry. A particularly noteworthy example is that of nitrilases, enzymes in which oligomerization of dimers into spiral homo-oligomers is a requirement for their enzymatic function. Nitrilases are widespread in nature where they catalyze the hydrolysis of nitriles into the corresponding carboxylic acid and ammonia. Here, we present the Cryo-EM structure, at 3 Å resolution, of a C-terminal truncate nitrilase from Rhodococcus sp. V51B that assembles in helical filaments. The model comprises a complete turn of the helical arrangement with a substrate-intermediate bound to the catalytic cysteine. The structure was solved having added the substrate to the protein. The length and stability of filaments was made more substantial in the presence of the aromatic substrate, benzonitrile, but not for aliphatic nitriles or dinitriles. The overall structure maintains the topology of the nitrilase family, and the filament is formed by the association of dimers in a chain-like mechanism that stabilizes the spiral. The active site is completely buried inside each monomer, while the substrate binding pocket was observed within the oligomerization interfaces. The present structure is in a closed configuration, judging by the position of the lid, suggesting that the intermediate is one of the covalent adducts. The proximity of the active site to the dimerization and oligomerization interfaces, allows the dimer to sense structural changes once the benzonitrile was bound, and translated to the rest of the filament, stabilizing the helical structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42779.map.gz emd_42779.map.gz | 163.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42779-v30.xml emd-42779-v30.xml emd-42779.xml emd-42779.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42779_fsc.xml emd_42779_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42779.png emd_42779.png | 78.1 KB | ||
| Masks |  emd_42779_msk_1.map emd_42779_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42779.cif.gz emd-42779.cif.gz | 6.3 KB | ||
| Others |  emd_42779_half_map_1.map.gz emd_42779_half_map_1.map.gz emd_42779_half_map_2.map.gz emd_42779_half_map_2.map.gz | 139 MB 138.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42779 http://ftp.pdbj.org/pub/emdb/structures/EMD-42779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42779 | HTTPS FTP |
-Validation report
| Summary document |  emd_42779_validation.pdf.gz emd_42779_validation.pdf.gz | 941.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42779_full_validation.pdf.gz emd_42779_full_validation.pdf.gz | 940.8 KB | Display | |
| Data in XML |  emd_42779_validation.xml.gz emd_42779_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_42779_validation.cif.gz emd_42779_validation.cif.gz | 26.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42779 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42779 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42779 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42779 | HTTPS FTP |
-Related structure data
| Related structure data |  8uxuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42779.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42779.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
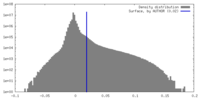
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42779_msk_1.map emd_42779_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
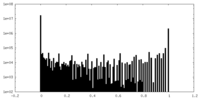
| Density Histograms |
-Half map: #2
| File | emd_42779_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
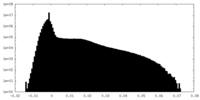
| Density Histograms |
-Half map: #1
| File | emd_42779_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
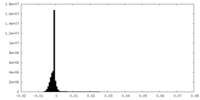
| Density Histograms |
- Sample components
Sample components
-Entire : Bacterial nitrilase filament with covalent adduct derived from be...
| Entire | Name: Bacterial nitrilase filament with covalent adduct derived from benzonitrile hydrolysis. |
|---|---|
| Components |
|
-Supramolecule #1: Bacterial nitrilase filament with covalent adduct derived from be...
| Supramolecule | Name: Bacterial nitrilase filament with covalent adduct derived from benzonitrile hydrolysis. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: C-terminal truncated mutant. |
|---|---|
| Source (natural) | Organism:  Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria) Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria)Strain: V51B |
-Macromolecule #1: Nitrilase
| Macromolecule | Name: Nitrilase / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria) Rhodococcus sp. (in: high G+C Gram-positive bacteria) (bacteria)Strain: V51B |
| Molecular weight | Theoretical: 36.25484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVEYTNTFKV AAVQAQPVWF DAAKTVDKTV SNIAEAARNG CELVAFPEVF IPGYPYHIWV DSPLAGMAKF AVRYHENSLT MDSPHVQRL LDAARDHNIA VVVGISERDG GSLYMTQLII DADGQLVARR RKLKPTHVER SVYGEGNGSD ISVYDMPFAR L GALNCWEH ...String: MVEYTNTFKV AAVQAQPVWF DAAKTVDKTV SNIAEAARNG CELVAFPEVF IPGYPYHIWV DSPLAGMAKF AVRYHENSLT MDSPHVQRL LDAARDHNIA VVVGISERDG GSLYMTQLII DADGQLVARR RKLKPTHVER SVYGEGNGSD ISVYDMPFAR L GALNCWEH FQTLTKYAMY SMHEQVHVAS WPGMSLYQPE VPAFGVDAQL TATRMYALEG QTFVVCTTQV VTPEAHEFFC EN EEQRKLI GRGGGFARII GPDGRDLATP LAEDEEGILY ADIDLSAITL AKQAADPVGH YSRPDVLSLN FNQRRTTPVN TPL STIHAT H UniProtKB: Nitrilase |
-Macromolecule #2: benzaldehyde
| Macromolecule | Name: benzaldehyde / type: ligand / ID: 2 / Number of copies: 14 / Formula: HBX |
|---|---|
| Molecular weight | Theoretical: 106.122 Da |
| Chemical component information |  ChemComp-HBX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 3.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
Details: Monobasic and dibasic potassium phosphate mixed at 7.8 pH. | |||||||||
| Grid | Model: Quantifoil / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Grids were glow-discharged for 60 sec before deposition of 3 ul sample, blotted for 4 s, and vitrified by plunging into liquid ethane and stored in a cryobox in liquid nitrogen.. | |||||||||
| Details | Purified protein was centrifuged and incubated with 100 mM benzonitrile for 15 min and applied to the grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 2358 / Average exposure time: 2.04081633 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
| Output model |  PDB-8uxu: |
-Atomic model buiding 2
| Refinement | Overall B value: 40 |
|---|---|
| Output model |  PDB-8uxu: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)