[English] 日本語
 Yorodumi
Yorodumi- EMDB-42774: Structure of Heterochromatin Protein 1 (HP1) alpha in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Heterochromatin Protein 1 (HP1) alpha in complex with an H2A.Z nucleosome | |||||||||
 Map data Map data | primary map, consensus refinement with mask | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | heterochromatin / nucleosome / chromatin / GENE REGULATION / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationchromocenter / histone H3K9me2/3 reader activity / nucleosomal DNA binding / : / histone deacetylase complex / Transcriptional Regulation by E2F6 / histone methyltransferase complex / ribonucleoprotein complex binding / site of DNA damage / RNA polymerase II core promoter sequence-specific DNA binding ...chromocenter / histone H3K9me2/3 reader activity / nucleosomal DNA binding / : / histone deacetylase complex / Transcriptional Regulation by E2F6 / histone methyltransferase complex / ribonucleoprotein complex binding / site of DNA damage / RNA polymerase II core promoter sequence-specific DNA binding / pericentric heterochromatin / heterochromatin / transcription repressor complex / SUMOylation of chromatin organization proteins / Regulation of endogenous retroelements by KRAB-ZFP proteins / cellular response to estradiol stimulus / euchromatin / kinetochore / histone deacetylase binding / structural constituent of chromatin / nuclear envelope / heterochromatin formation / nucleosome / nucleosome assembly / Factors involved in megakaryocyte development and platelet production / chromatin organization / protein-macromolecule adaptor activity / DNA-binding transcription factor binding / chromosome, telomeric region / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / ribonucleoprotein complex / negative regulation of DNA-templated transcription / DNA damage response / chromatin binding / protein-containing complex binding / nucleolus / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Tan D / Sokolova V | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Structural mechanism of HP1⍺-dependent transcriptional repression and chromatin compaction. Authors: Vladyslava Sokolova / Jacob Miratsky / Vladimir Svetlov / Michael Brenowitz / John Vant / Tyler S Lewis / Kelly Dryden / Gahyun Lee / Shayan Sarkar / Evgeny Nudler / Abhishek Singharoy / Dongyan Tan /  Abstract: Heterochromatin protein 1 (HP1) plays a central role in establishing and maintaining constitutive heterochromatin. However, the mechanisms underlying HP1-nucleosome interactions and their ...Heterochromatin protein 1 (HP1) plays a central role in establishing and maintaining constitutive heterochromatin. However, the mechanisms underlying HP1-nucleosome interactions and their contributions to heterochromatin functions remain elusive. Here, we present the cryoelectron microscopy (cryo-EM) structure of an HP1α dimer bound to an H2A.Z-nucleosome, revealing two distinct HP1α-nucleosome interfaces. The primary HP1α binding site is located at the N terminus of histone H3, specifically at the trimethylated lysine 9 (K9me3) region, while a secondary binding site is situated near histone H2B, close to nucleosome superhelical location 4 (SHL4). Our biochemical data further demonstrates that HP1α binding influences the dynamics of DNA on the nucleosome. It promotes DNA unwrapping near the nucleosome entry and exit sites while concurrently restricting DNA accessibility in the vicinity of SHL4. Our study offers a model for HP1α-mediated heterochromatin maintenance and gene silencing. It also sheds light on the H3K9me-independent role of HP1 in responding to DNA damage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42774.map.gz emd_42774.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42774-v30.xml emd-42774-v30.xml emd-42774.xml emd-42774.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42774.png emd_42774.png | 55.2 KB | ||
| Filedesc metadata |  emd-42774.cif.gz emd-42774.cif.gz | 7.6 KB | ||
| Others |  emd_42774_half_map_1.map.gz emd_42774_half_map_1.map.gz emd_42774_half_map_2.map.gz emd_42774_half_map_2.map.gz | 49.8 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42774 http://ftp.pdbj.org/pub/emdb/structures/EMD-42774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42774 | HTTPS FTP |
-Related structure data
| Related structure data |  8uxqMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42774.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42774.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map, consensus refinement with mask | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
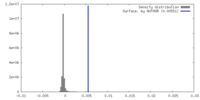
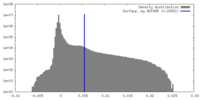
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : HP1alpha dimer bound to H2A.Z nucleosome
| Entire | Name: HP1alpha dimer bound to H2A.Z nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: HP1alpha dimer bound to H2A.Z nucleosome
| Supramolecule | Name: HP1alpha dimer bound to H2A.Z nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 288 KDa |
-Macromolecule #1: Chromobox protein homolog 5
| Macromolecule | Name: Chromobox protein homolog 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.651398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSPEFGKKTK RTADSSSSED EEEYVVEKVL DRRVVKGQVE YLLKWKGFSE EHNTWEPEKN LDCPELISEF MKKYKKMKEG ENNKPREKS ESNKRKSNFS NSADDIKSKK KREQSNDIAR GFERGLEPEK IIGATDSCGD LMFLMKWKDT DEADLVLAKE A NVKCPQIV ...String: GSPEFGKKTK RTADSSSSED EEEYVVEKVL DRRVVKGQVE YLLKWKGFSE EHNTWEPEKN LDCPELISEF MKKYKKMKEG ENNKPREKS ESNKRKSNFS NSADDIKSKK KREQSNDIAR GFERGLEPEK IIGATDSCGD LMFLMKWKDT DEADLVLAKE A NVKCPQIV IAFYEERLTW HAYPEDAENK EKETAKS UniProtKB: Chromobox protein homolog 5 |
-Macromolecule #2: Histone H3.2
| Macromolecule | Name: Histone H3.2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.30393 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3.2 |
-Macromolecule #3: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.263231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #4: Histone H2A.Z
| Macromolecule | Name: Histone H2A.Z / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.450601 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AGGKAGKDSG KAKTKAVSRS QRAGLQFPVG RIHRHLKSRT TSHGRVGATA AVYSAAILEY LTAEVLELAG NASKDLKVKR ITPRHLQLA IRGDEELDSL IKATIAGGGV IPHIHKSLIG KKGQQKTV UniProtKB: Histone H2A.Z |
-Macromolecule #5: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.848097 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PEPAKSAPAP KKGSKKAVTK TQKKDGKKRR KTRKESYAIY VYKVLKQVHP DTGISSKAMS IMNSFVNDVF ERIAGEASRL AHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #6: DNA Widom601 (208bp) strand1
| Macromolecule | Name: DNA Widom601 (208bp) strand1 / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 64.456039 KDa |
| Sequence | String: (DA)(DC)(DT)(DC)(DC)(DG)(DG)(DC)(DA)(DA) (DG)(DG)(DT)(DC)(DG)(DC)(DT)(DG)(DT)(DT) (DC)(DA)(DA)(DT)(DA)(DC)(DA)(DT)(DG) (DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT) (DA) (DT)(DA)(DT)(DA)(DT)(DC) ...String: (DA)(DC)(DT)(DC)(DC)(DG)(DG)(DC)(DA)(DA) (DG)(DG)(DT)(DC)(DG)(DC)(DT)(DG)(DT)(DT) (DC)(DA)(DA)(DT)(DA)(DC)(DA)(DT)(DG) (DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT) (DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG) (DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT) (DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT)(DC)(DC) (DC)(DC)(DT) (DT)(DG)(DG)(DC)(DG)(DG) (DT)(DT)(DA)(DA)(DA)(DA)(DC)(DG)(DC)(DG) (DG)(DG)(DG)(DG) (DA)(DC)(DA)(DG)(DC) (DG)(DC)(DG)(DT)(DA)(DC)(DG)(DT)(DG)(DC) (DG)(DT)(DT)(DT)(DA) (DA)(DG)(DC)(DG) (DG)(DT)(DG)(DC)(DT)(DA)(DG)(DA)(DG)(DC) (DT)(DG)(DT)(DC)(DT)(DA) (DC)(DG)(DA) (DC)(DC)(DA)(DA)(DT)(DT)(DG)(DA)(DG)(DC) (DG)(DG)(DC)(DC)(DT)(DC)(DG) (DG)(DC) (DA)(DC)(DC)(DG)(DG)(DG)(DA)(DT)(DT)(DC) (DT)(DC)(DC)(DA)(DG)(DG)(DG)(DC) (DG) (DG)(DC)(DC)(DG)(DC)(DG)(DT)(DA)(DT)(DA) (DG)(DG)(DG)(DT)(DC)(DC)(DA)(DT)(DC) (DA)(DC)(DA)(DT)(DA)(DA)(DG)(DT) |
-Macromolecule #7: DNA Widom601 (208bp) strand2
| Macromolecule | Name: DNA Widom601 (208bp) strand2 / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 63.988711 KDa |
| Sequence | String: (DA)(DC)(DT)(DT)(DA)(DT)(DG)(DT)(DG)(DA) (DT)(DG)(DG)(DA)(DC)(DC)(DC)(DT)(DA)(DT) (DA)(DC)(DG)(DC)(DG)(DG)(DC)(DC)(DG) (DC)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA) (DT) (DC)(DC)(DC)(DG)(DG)(DT) ...String: (DA)(DC)(DT)(DT)(DA)(DT)(DG)(DT)(DG)(DA) (DT)(DG)(DG)(DA)(DC)(DC)(DC)(DT)(DA)(DT) (DA)(DC)(DG)(DC)(DG)(DG)(DC)(DC)(DG) (DC)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA) (DT) (DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC)(DT) (DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG)(DT)(DA)(DG)(DA)(DC)(DA)(DG)(DC)(DT) (DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG) (DC)(DT)(DT)(DA)(DA)(DA)(DC)(DG)(DC)(DA) (DC)(DG)(DT)(DA) (DC)(DG)(DC)(DG)(DC) (DT)(DG)(DT)(DC)(DC)(DC)(DC)(DC)(DG)(DC) (DG)(DT)(DT)(DT)(DT) (DA)(DA)(DC)(DC) (DG)(DC)(DC)(DA)(DA)(DG)(DG)(DG)(DG)(DA) (DT)(DT)(DA)(DC)(DT)(DC) (DC)(DC)(DT) (DA)(DG)(DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG) (DC)(DA)(DC)(DG)(DT)(DG)(DT) (DC)(DA) (DG)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DC)(DT)(DG)(DT)(DG)(DC) (DA) (DT)(DG)(DT)(DA)(DT)(DT)(DG)(DA)(DA)(DC) (DA)(DG)(DC)(DG)(DA)(DC)(DC)(DT)(DT) (DG)(DC)(DC)(DG)(DG)(DA)(DG)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 39.2 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)