[English] 日本語
 Yorodumi
Yorodumi- EMDB-4258: Structure of Ryanodine receptor 1 in nanodiscs in the presence of... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4258 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Ryanodine receptor 1 in nanodiscs in the presence of calcium, ATP and ryanodine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calcium release channel / nanodiscs / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity ...ATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / outflow tract morphogenesis / toxic substance binding / striated muscle contraction / voltage-gated calcium channel activity / skeletal muscle fiber development / release of sequestered calcium ion into cytosol / sarcoplasmic reticulum membrane / cellular response to calcium ion / muscle contraction / sarcoplasmic reticulum / sarcolemma / calcium ion transmembrane transport / calcium channel activity / Z disc / intracellular calcium ion homeostasis / disordered domain specific binding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / calcium ion binding / ATP binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
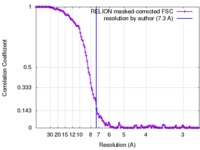
| Method | single particle reconstruction / cryo EM / Resolution: 7.3 Å | |||||||||
 Authors Authors | Willegems K / Efremov RG | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: Influence of Lipid Mimetics on Gating of Ryanodine Receptor. Authors: Katrien Willegems / Rouslan G Efremov /  Abstract: Understanding gating principles of ion channels at high resolution is of great importance. Here we investigate the conformational transition from closed to open state in ryanodine receptor 1 (RyR1) ...Understanding gating principles of ion channels at high resolution is of great importance. Here we investigate the conformational transition from closed to open state in ryanodine receptor 1 (RyR1) reconstituted into lipid nanodiscs. RyR1 is a homotetrameric giant ion channel that couples excitation of muscle cells to fast calcium release from the sarcoplasmic reticulum. Using single-particle cryo-EM we show that RyR1 reconstituted into lipid nanodiscs is stabilized in the open conformation when bound to the plant toxin ryanodine, but not in the presence of its physiological activators, calcium and ATP. Further, using ryanodine binding assays we show that membrane mimetics influence RyR1 transition between closed and open-channel conformations. We find that all detergents, including fluorinated detergents added to nanodiscs, stabilize closed state of RyR1. Our biochemical results correlate with available structural data and suggest optimal conditions for structural studies of RyR1 gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4258.map.gz emd_4258.map.gz | 195.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4258-v30.xml emd-4258-v30.xml emd-4258.xml emd-4258.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4258_fsc.xml emd_4258_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_4258.png emd_4258.png | 99.8 KB | ||
| Filedesc metadata |  emd-4258.cif.gz emd-4258.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4258 http://ftp.pdbj.org/pub/emdb/structures/EMD-4258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4258 | HTTPS FTP |
-Related structure data
| Related structure data |  6fg3MC  4295C  6fooC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4258.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4258.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.355 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ryanodine receptor 1 reconstituted in lipid nanodiscs in the pres...
| Entire | Name: Ryanodine receptor 1 reconstituted in lipid nanodiscs in the presence of calcium, ATP and ryanodine |
|---|---|
| Components |
|
-Supramolecule #1: Ryanodine receptor 1 reconstituted in lipid nanodiscs in the pres...
| Supramolecule | Name: Ryanodine receptor 1 reconstituted in lipid nanodiscs in the presence of calcium, ATP and ryanodine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.2 MDa |
-Macromolecule #1: Ryanodine receptor 1
| Macromolecule | Name: Ryanodine receptor 1 / type: protein_or_peptide / ID: 1 Details: Poly Unk stretches correspond to polypeptide regions where the register is not assigned. Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 532.033062 KDa |
| Sequence | String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD ...String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD LILVSVSSER YLHLSTASGE LQVDASFMQT LWNMNPICSC CEEGYVTGGH VLRLFHGHMD ECLTISAADS DD QRRLVYY EGGAVCTHAR SLWRLEPLRI SWSGSHLRWG QPLRIRHVTT GRYLALTEDQ GLVVVDACKA HTKATSFCFR VSK EKLDTA PKRDVEGMGP PEIKYGESLC FVQHVASGLW LTYAAPDPKA LRLGVLKKKA ILHQEGHMDD ALFLTRCQQE ESQA ARMIH STAGLYNQFI KGLDSFSGKP RGSGPPAGPA LPIEAVILSL QDLIGYFEPP SEELQHEEKQ SKLRSLRNRQ SLFQE EGML SLVLNCIDRL NVYTTAAHFA EYAGEEAAES WKEIVNLLYE LLASLIRGNR ANCALFSTNL DWVVSKLDRL EASSGI LEV LYCVLIESPE VLNIIQENHI KSIISLLDKH GRNHKVLDVL CSLCVCNGVA VRSNQDLITE NLLPGRELLL QTNLINY VT SIRPNIFVGR AEGSTQYGKW YFEVMVDEVV PFLTAQATHL RVGWALTEGY SPYPGGGEGW GGNGVGDDLY SYGFDGLH L WTGHVARPVT SPGQHLLAPE DVVSCCLDLS VPSISFRING CPVQGVFEAF NLDGLFFPVV SFSAGVKVRF LLGGRHGEF KFLPPPGYAP CHEAVLPRER LRLEPIKEYR REGPRGPHLV GPSRCLSHTD FVPCPVDTVQ IVLPPHLERI REKLAENIHE LWALTRIEQ GWTYGPVRDD NKRLHPCLVN FHSLPEPERN YNLQMSGETL KTLLALGCHV GMADEKAEDN LKKTKLPKTY M MSNGYKPA PLDLSHVRLT PAQTTLVDRL AENGHNVWAR DRVAQGWSYS AVQDIPARRN PRLVPYRLLD EATKRSNRDS LC QAVRTLL GYGYNIEPPD QEPSQVENQS RWDRVRIFRA EKSYTVQSGR WYFEFEAVTT GEMRVGWARP ELRPDVELGA DEL AYVFNG HRGQRWHLGS EPFGRPWQSG DVVGCMIDLT ENTIIFTLNG EVLMSDSGSE TAFREIEIGD GFLPVCSLGP GQVG HLNLG QDVSSLRFFA ICGLQEGFEP FAINMQRPVT TWFSKSLPQF EPVPPEHPHY EVARMDGTVD TPPCLRLAHR (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)MPLS AAMFLSERKN PAPQCPPRLE VQMLMPVSWS RMPNHF LQV ETRRAGERLG WAVQCQDPLT MMALHIPEEN RCMDILELSE RLDLQRFHSH TLRLYRAVCA LGNNRVAHAL CSHVDQA QL LHALEDAHLP GPLRAGYYDL LISIHLESAC RSRRSMLSEY IVPLTPETRA ITLFPPGRKG GNARRHGLPG VGVTTSLR P PHHFSPPCFV AALPAAGVAE APARLSPAIP LEALRDKALR MLGEAVRDGG QHARDPVGGS VEFQFVPVLK LVSTLLVMG IFGDEDVKQI LKMIEPEVFT EEEEEEEEEE EEEEEEEEDE EEKEEDEEEE EKEDAEKEEE EAPEGEKEDL EEGLLQMKLP ESVKLQMCN LLEYFCDQEL QHRVESLAAF AERYVDKLQA NQRSRYALLM RAFTMSAAET ARRTREFRSP PQEQINMLLH F KDEADEED CPLPEDIRQD LQDFHQDLLA HCGIQLEGEE EEPEEETSLS SRLRSLLETV RLVKKKEEKP EEELPAEEKK PQ SLQELVS HMVVRWAQED YVQSPELVRA MFSLLHRQYD GLGELLRALP RAYTISPSSV EDTMSLLECL GQIRSLLIVQ MGP QEENLM IQSIGNIMNN KVFYQHPNLM RALGMHETVM EVMVNVLGGG ETKEIRFPKM VTSCCRFLCY FCRISRQNQR SMFD HLSYL LENSGIGLGM QGSTPLDVAA ASVIDNNELA LALQEQDLEK VVSYLAGCGL QSCPMLLAKG YPDIGWNPCG GERYL DFLR FAVFVNGESV EENANVVVRL LIRKPECFGP ALRGEGGSGL LAAIEEAIRI SEDPARDGPG VRRDRRREHF GEEPPE ENR VHLGHAIMSF YAALIDLLGR CAPEMHLIQA GKGEALRIRA ILRSLVPLDD LVGIISLPLQ IPTL(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)NFDPRP VETLNVIIPE KLDS FINKF AEYTHEKWAF DKIQNNWSYG ENVDEELKTH PMLRPYKTFS EKDKEIYRWP IKESLKAMIA WEWTIEKARE GEEER TEKK KTRKISQTAQ TYDPREGYNP QPPDLSGVTL SRELQAMAEQ LAENYHNTWG RKKKQELEAK GGGTHPLLVP YDTLTA KEK ARDREKAQEL LKFLQMNGYA VTR(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)TPLY NLPTHRACNM FLESYKAAWI LTEDHSFEDR MIDDLSKAGE QEEEEEEVEE KKPDPLHQLV LHFSRTALTE K SKLDEDYL YMAYADIMAK SCHLEEGGEN GEAEEEEVEV SFEEKEMEKQ RLLYQQSRLH TRGAAEMVLQ MISACKGETG AM VSSTLKL GISILNGGNA EVQQKMLDYL KDKKEVGFFQ SIQALMQTCS VLDLNAFERQ NKAEGLGMVN EDGTVINRQN GEK VMADDE FTQDLFRFLQ LLCEGHNNDF QNYLRTQTGN TTTINIIICT VDYLLRLQES ISDFYWYYSG KDVIEEQGKR NFSK AMSVA KQVFNSLTEY IQGPCTGNQQ SLAHSRLWDA VVGFLHVFAH MMMKLAQDSS QIELLKELLD LQKDMVVMLL SLLEG NVVN GMIARQMVDM LVESSSNVEM ILKFFDMFLK LKDIVGSEAF QDYVTDPRGL ISKKDFQKAM DSQKQFTGPE IQFLLS CSE ADENEMINFE EFANRFQEPA RDIGFNVAVL LTNLSEHVPH DPRLRNFLEL AESILEYFRP YLGRIEIMGA SRRIERI YF EISETNRAQW EMPQVKESKR QFIFDVVNEG GEAEKMELFV SFCEDTIFEM QIAAQISEPE GEPEADEDEG MGEAAAEG A EEGAAGAEGA AGTVAAGATA RLAAAAARAL RGLSYRSLRR RVRRLRRLTA REAATALAAL LWAVVARAGA AGAGAAAGA LRLLWGSLFG GGLVEGAKKV TVTELLAGMP DPTSDEVHGE QPAGPGGDAD GAGEGEGEGD AAEGDGDEEV AGHEAGPGGA EGVVAVADG GPFRPEGAGG LGDMGDTTPA EPPTPEGSPI LKRKLGVDGE EEELVPEPEP EPEPEPEKAD EENGEKEEVP E APPEPPKK APPSPPAKKE EAGGAGMEFW GELEVQRVKF LNYLSRNFYT LRFLALFLAF AINFILLFYK VSDSPPGEDD ME GSAAGDL AGAGSGGGSG WGSGAGEEAE GDEDENMVYY FLEESTGYME PALWCLSLLH TLVAFLCIIG YNCLKVPLVI FKR EKELAR KLEFDGLYIT EQPGDDDVKG QWDRLVLNTP SFPSNYWDKF VKRKVLDKHG DIFGRERIAE LLGMDLASLE ITAH NERKP DPPPGLLTWL MSIDVKYQIW KFGVIFTDNS FLYLGWYMVM SLLGHYNNFF FAAHLLDIAM GVKTLRTILS SVTHN GKQL VMTVGLLAVV VYLYTVVAFN FFRKFYNKSE DEDEPDMKCD DMMTCYLFHM YVGVRAGGGI GDEIEDPAGD EYELYR VVF DITFFFFVIV ILLAIIQGLI IDAFGELRDQ QEQVKEDMET KCFICGIGSD YFDTTPHGFE THTLEEHNLA NYMFFLM YL INKDETEHTG QESYVWKMYQ ERCWDFFPAG DCFRKQYEDQ LS UniProtKB: Ryanodine receptor 1, Ryanodine receptor 1, Ryanodine receptor 1, Ryanodine receptor 1 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.04 kPa Details: Glow discharged in ELMO plasma cleaner at current of 3 mA for 1 min. | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 293.15 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-7 / Number grids imaged: 1 / Number real images: 3078 / Average exposure time: 1.0 sec. / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6fg3: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)