+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of synaptic vesicle protein 2B with padsevonil | |||||||||
 Map data Map data | Main map sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic vesicle / SLC22 / Inhibitor / Antiepileptic / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationToxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / regulation of presynaptic cytosolic calcium ion concentration / neurotransmitter transport / regulation of synaptic vesicle exocytosis / transmembrane transporter activity / acrosomal vesicle / synaptic vesicle ...Toxicity of botulinum toxin type F (botF) / Toxicity of botulinum toxin type D (botD) / Toxicity of botulinum toxin type E (botE) / Toxicity of botulinum toxin type A (botA) / regulation of presynaptic cytosolic calcium ion concentration / neurotransmitter transport / regulation of synaptic vesicle exocytosis / transmembrane transporter activity / acrosomal vesicle / synaptic vesicle / synaptic vesicle membrane / chemical synaptic transmission / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
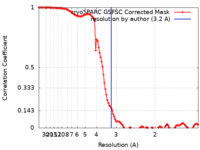
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Martin MF / Mittal A / Levin E / Adams C / Yang M / Ledecq M / Horanyi PS / Coleman JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants. Authors: Anshumali Mittal / Matthew F Martin / Elena J Levin / Christopher Adams / Meng Yang / Laurent Provins / Adrian Hall / Martin Procter / Marie Ledecq / Alexander Hillisch / Christian Wolff / ...Authors: Anshumali Mittal / Matthew F Martin / Elena J Levin / Christopher Adams / Meng Yang / Laurent Provins / Adrian Hall / Martin Procter / Marie Ledecq / Alexander Hillisch / Christian Wolff / Michel Gillard / Peter S Horanyi / Jonathan A Coleman /     Abstract: Epilepsy is a common neurological disorder characterized by abnormal activity of neuronal networks, leading to seizures. The racetam class of anti-seizure medications bind specifically to a membrane ...Epilepsy is a common neurological disorder characterized by abnormal activity of neuronal networks, leading to seizures. The racetam class of anti-seizure medications bind specifically to a membrane protein found in the synaptic vesicles of neurons called synaptic vesicle protein 2 (SV2) A (SV2A). SV2A belongs to an orphan subfamily of the solute carrier 22 organic ion transporter family that also includes SV2B and SV2C. The molecular basis for how anti-seizure medications act on SV2s remains unknown. Here we report cryo-electron microscopy structures of SV2A and SV2B captured in a luminal-occluded conformation complexed with anticonvulsant ligands. The conformation bound by anticonvulsants resembles an inhibited transporter with closed luminal and intracellular gates. Anticonvulsants bind to a highly conserved central site in SV2s. These structures provide blueprints for future drug design and will facilitate future investigations into the biological function of SV2s. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42430.map.gz emd_42430.map.gz | 136.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42430-v30.xml emd-42430-v30.xml emd-42430.xml emd-42430.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42430_fsc.xml emd_42430_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_42430.png emd_42430.png | 36 KB | ||
| Masks |  emd_42430_msk_1.map emd_42430_msk_1.map emd_42430_msk_2.map emd_42430_msk_2.map | 144.7 MB 144.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42430.cif.gz emd-42430.cif.gz | 7.1 KB | ||
| Others |  emd_42430_additional_1.map.gz emd_42430_additional_1.map.gz emd_42430_half_map_1.map.gz emd_42430_half_map_1.map.gz emd_42430_half_map_2.map.gz emd_42430_half_map_2.map.gz | 72.2 MB 134.1 MB 134.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42430 http://ftp.pdbj.org/pub/emdb/structures/EMD-42430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42430 | HTTPS FTP |
-Related structure data
| Related structure data |  8uo8MC  8uo9C  8uoaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42430.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42430.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.72 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42430_msk_1.map emd_42430_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
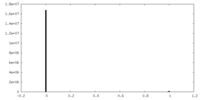
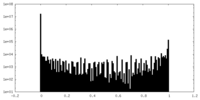
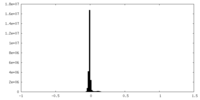
| Density Histograms |
-Mask #2
| File |  emd_42430_msk_2.map emd_42430_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
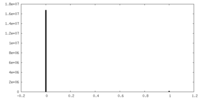
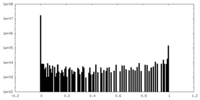
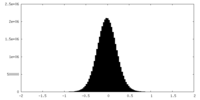
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_42430_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
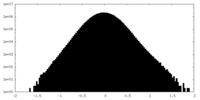
| Density Histograms |
-Half map: Half map B
| File | emd_42430_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
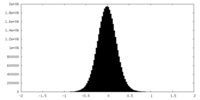
| Density Histograms |
-Half map: Half map A
| File | emd_42430_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SV2B complexed with padsevonil
| Entire | Name: SV2B complexed with padsevonil |
|---|---|
| Components |
|
-Supramolecule #1: SV2B complexed with padsevonil
| Supramolecule | Name: SV2B complexed with padsevonil / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.4 kDa/nm |
-Macromolecule #1: Synaptic vesicle glycoprotein 2B
| Macromolecule | Name: Synaptic vesicle glycoprotein 2B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.515016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDDYKYQDNY GGYAPSDGYY RGNESNPEED AQSDVTEGHD EEDEIYEGEY QGIPHPDDVK AKQAKMAPSR MDSLRGQTDL MAERLEDEE QLAHQYETIM DECGHGRFQW ILFFVLGLAL MADGVEVFVV SFALPSAEKD MCLSSSKKGM LGMIVYLGMM A GAFILGGL ...String: MDDYKYQDNY GGYAPSDGYY RGNESNPEED AQSDVTEGHD EEDEIYEGEY QGIPHPDDVK AKQAKMAPSR MDSLRGQTDL MAERLEDEE QLAHQYETIM DECGHGRFQW ILFFVLGLAL MADGVEVFVV SFALPSAEKD MCLSSSKKGM LGMIVYLGMM A GAFILGGL ADKLGRKRVL SMSLAVNASF ASLSSFVQGY GAFLFCRLIS GIGIGGALPI VFAYFSEFLS REKRGEHLSW LG IFWMTGG LYASAMAWSI IPHYGWGFSM GTNYHFHSWR VFVIVCALPC TVSMVALKFM PESPRFLLEM GKHDEAWMIL KQV HDTNMR AKGTPEKVFT VSNIKTPKQM DEFIEIQSST GTWYQRWLVR FKTIFKQVWD NALYCVMGPY RMNTLILAVV WFAM AFSYY GLTVWFPDMI RYFQDEEYKS KMKVFFGEHV YGATINFTME NQIHQHGKLV NDKFTRMYFK HVLFEDTFFD ECYFE DVTS TDTYFKNCTI ESTIFYNTDL YEHKFINCRF INSTFLEQKE GCHMDLEQDN DFLIYLVSFL GSLSVLPGNI ISALLM DRI GRLKMIGGSM LISAVCCFFL FFGNSESAMI GWQCLFCGTS IAAWNALDVI TVELYPTNQR ATAFGILNGL CKFGAIL GN TIFASFVGIT KVVPILLAAA SLVGGGLIAL RLPETREQVL M UniProtKB: Synaptic vesicle glycoprotein 2B |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: (4R)-4-(2-chloro-2,2-difluoroethyl)-1-{[(4R)-2-(methoxymethyl)-6-...
| Macromolecule | Name: (4R)-4-(2-chloro-2,2-difluoroethyl)-1-{[(4R)-2-(methoxymethyl)-6-(trifluoromethyl)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]methyl}pyrrolidin-2-one type: ligand / ID: 3 / Number of copies: 1 / Formula: X3U |
|---|---|
| Molecular weight | Theoretical: 432.797 Da |
-Macromolecule #4: 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE]
| Macromolecule | Name: 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE] / type: ligand / ID: 4 / Number of copies: 1 / Formula: PS1 |
|---|---|
| Molecular weight | Theoretical: 566.642 Da |
| Chemical component information |  ChemComp-PS1: |
-Macromolecule #5: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 5 / Number of copies: 2 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Macromolecule #6: [(2R)-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]o...
| Macromolecule | Name: [(2R)-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-2-propanoyloxy-propyl] propanoate type: ligand / ID: 6 / Number of copies: 1 / Formula: 43Y |
|---|---|
| Molecular weight | Theoretical: 370.356 Da |
| Chemical component information | 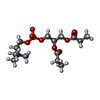 ChemComp-43Y: |
-Macromolecule #7: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 150 mM NaCl, 20 mM Tris pH 8.0, .4 mM glyco-diosgenin, 1 uM padsevonil |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 194000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)