+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4215 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast 80S ribosome with an uncleaved 20S rRNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Scaiola A / Pena C / Weisser M / Boehringer D / Leibundgut M / Klingauf-Nerurkar P / Gerhardy S / Panse VG / Ban N | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2018 Journal: EMBO J / Year: 2018Title: Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. Authors: Alain Scaiola / Cohue Peña / Melanie Weisser / Daniel Böhringer / Marc Leibundgut / Purnima Klingauf-Nerurkar / Stefan Gerhardy / Vikram Govind Panse / Nenad Ban /  Abstract: Final maturation of eukaryotic ribosomes occurs in the cytoplasm and requires the sequential removal of associated assembly factors and processing of the immature 20S pre-RNA Using cryo-electron ...Final maturation of eukaryotic ribosomes occurs in the cytoplasm and requires the sequential removal of associated assembly factors and processing of the immature 20S pre-RNA Using cryo-electron microscopy (cryo-EM), we have determined the structure of a yeast cytoplasmic pre-40S particle in complex with Enp1, Ltv1, Rio2, Tsr1, and Pno1 assembly factors poised to initiate final maturation. The structure reveals that the pre-rRNA adopts a highly distorted conformation of its 3' major and 3' minor domains stabilized by the binding of the assembly factors. This observation is consistent with a mechanism that involves concerted release of the assembly factors orchestrated by the folding of the rRNA in the head of the pre-40S subunit during the final stages of maturation. Our results provide a structural framework for the coordination of the final maturation events that drive a pre-40S particle toward the mature form capable of engaging in translation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4215.map.gz emd_4215.map.gz | 18.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4215-v30.xml emd-4215-v30.xml emd-4215.xml emd-4215.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4215.png emd_4215.png | 86 KB | ||
| Others |  emd_4215_half_map_1.map.gz emd_4215_half_map_1.map.gz emd_4215_half_map_2.map.gz emd_4215_half_map_2.map.gz | 98.1 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4215 http://ftp.pdbj.org/pub/emdb/structures/EMD-4215 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4215 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4215 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4215.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4215.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
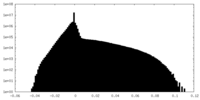
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_4215_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
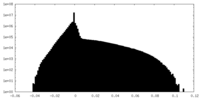
| Density Histograms |
-Half map: #2
| File | emd_4215_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
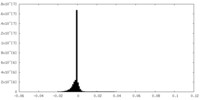
| Density Histograms |
- Sample components
Sample components
-Entire : Cytoplasmic yeast 80S ribosome with a uncleaved 20S rRNA
| Entire | Name: Cytoplasmic yeast 80S ribosome with a uncleaved 20S rRNA |
|---|---|
| Components |
|
-Supramolecule #1: Cytoplasmic yeast 80S ribosome with a uncleaved 20S rRNA
| Supramolecule | Name: Cytoplasmic yeast 80S ribosome with a uncleaved 20S rRNA type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2) / Number images used: 145081 |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)