[English] 日本語
 Yorodumi
Yorodumi- EMDB-41845: Gaussian mixture model based single particle refinement - ABC tra... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
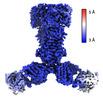
| Title | Gaussian mixture model based single particle refinement - ABC transporter (inhibitor-bound ABCG2 from EMPIAR-10374) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbiotin transmembrane transporter activity / biotin transport / riboflavin transport / riboflavin transmembrane transporter activity / sphingolipid transporter activity / renal urate salt excretion / Abacavir transmembrane transport / urate metabolic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis ...biotin transmembrane transporter activity / biotin transport / riboflavin transport / riboflavin transmembrane transporter activity / sphingolipid transporter activity / renal urate salt excretion / Abacavir transmembrane transport / urate metabolic process / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / urate transmembrane transporter activity / external side of apical plasma membrane / xenobiotic transport across blood-brain barrier / organic anion transport / : / transepithelial transport / Ciprofloxacin ADME / Paracetamol ADME / export across plasma membrane / NFE2L2 regulating MDR associated enzymes / Differentiation of Keratinocytes in Interfollicular Epidermis in Mammalian Skin / ABC-type xenobiotic transporter / Heme biosynthesis / cellular detoxification / ABC-type xenobiotic transporter activity / Heme degradation / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / transport across blood-brain barrier / brush border membrane / Iron uptake and transport / transmembrane transport / mitochondrial membrane / apical plasma membrane / membrane raft / protein homodimerization activity / ATP hydrolysis activity / nucleoplasm / ATP binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Chen M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2024 Journal: Nat Methods / Year: 2024Title: Improving resolution and resolvability of single-particle cryoEM structures using Gaussian mixture models. Authors: Muyuan Chen / Michael F Schmid / Wah Chiu /  Abstract: Cryogenic electron microscopy is widely used in structural biology, but its resolution is often limited by the dynamics of the macromolecule. Here we developed a refinement protocol based on Gaussian ...Cryogenic electron microscopy is widely used in structural biology, but its resolution is often limited by the dynamics of the macromolecule. Here we developed a refinement protocol based on Gaussian mixture models that integrates particle orientation and conformation estimation and improves the alignment for flexible domains of protein structures. We demonstrated this protocol on multiple datasets, resulting in improved resolution and resolvability, locally and globally, by visual and quantitative measures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41845.map.gz emd_41845.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41845-v30.xml emd-41845-v30.xml emd-41845.xml emd-41845.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41845_fsc.xml emd_41845_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41845.png emd_41845.png | 130.8 KB | ||
| Filedesc metadata |  emd-41845.cif.gz emd-41845.cif.gz | 6.6 KB | ||
| Others |  emd_41845_half_map_1.map.gz emd_41845_half_map_1.map.gz emd_41845_half_map_2.map.gz emd_41845_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41845 http://ftp.pdbj.org/pub/emdb/structures/EMD-41845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41845 | HTTPS FTP |
-Related structure data
| Related structure data |  8u2cMC  8u26C  8u28C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41845.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41845.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41845_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41845_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : inhibitor-bound ABCG2
| Entire | Name: inhibitor-bound ABCG2 |
|---|---|
| Components |
|
-Supramolecule #1: inhibitor-bound ABCG2
| Supramolecule | Name: inhibitor-bound ABCG2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Re-refinement from EMPIAR-10374 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Broad substrate specificity ATP-binding cassette transporter ABCG2
| Macromolecule | Name: Broad substrate specificity ATP-binding cassette transporter ABCG2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.385852 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSSNVEVFI PVSQGNTNGF PATASNDLKA FTEGAVLSFH NICYRVKLKS GFLPCRKPVE KEILSNINGI MKPGLNAILG PTGGGKSSL LDVLAARKDP SGLSGDVLIN GAPRPANFKC NSGYVVQDDV VMGTLTVREN LQFSAALRLA TTMTNHEKNE R INRVIQEL ...String: MSSSNVEVFI PVSQGNTNGF PATASNDLKA FTEGAVLSFH NICYRVKLKS GFLPCRKPVE KEILSNINGI MKPGLNAILG PTGGGKSSL LDVLAARKDP SGLSGDVLIN GAPRPANFKC NSGYVVQDDV VMGTLTVREN LQFSAALRLA TTMTNHEKNE R INRVIQEL GLDKVADSKV GTQFIRGVSG GERKRTSIGM ELITDPSILF LDEPTTGLDS STANAVLLLL KRMSKQGRTI IF SIHQPRY SIFKLFDSLT LLASGRLMFH GPAQEALGYF ESAGYHCEAY NNPADFFLDI INGDSTAVAL NREEDFKATE IIE PSKQDK PLIEKLAEIY VNSSFYKETK AELHQLSGGE KKKKITVFKE ISYTTSFCHQ LRWVSKRSFK NLLGNPQASI AQII VTVVL GLVIGAIYFG LKNDSTGIQN RAGVLFFLTT NQCFSSVSAV ELFVVEKKLF IHEYISGYYR VSSYFLGKLL SDLLP MRML PSIIFTCIVY FMLGLKPKAD AFFVMMFTLM MVAYSASSMA LAIAAGQSVV SVATLLMTIC FVFMMIFSGL LVNLTT IAS WLSWLQYFSI PRYGFTALQH NEFLGQNFCP GLNATGNNPC NYATCTGEEY LVKQGIDLSP WGLWKNHVAL ACMIVIF LT IAYLKLLFLK KYS UniProtKB: Broad substrate specificity ATP-binding cassette transporter ABCG2 |
-Macromolecule #2: 5D3 Fab light chain variable domain
| Macromolecule | Name: 5D3 Fab light chain variable domain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.594016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVLTQSPSS FSVSLGDRVT ISCKASGYIL NRLAWYQQKP GNAPRLLISG ATSLETGFPS RFSGTGSGKD YTLSISSLQT EDVGTYYCQ QYWSTPWTFG GGTKLEIRRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVLTQSPSS FSVSLGDRVT ISCKASGYIL NRLAWYQQKP GNAPRLLISG ATSLETGFPS RFSGTGSGKD YTLSISSLQT EDVGTYYCQ QYWSTPWTFG GGTKLEIRRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Macromolecule #3: 5D3 Fab heavy chain variable domain
| Macromolecule | Name: 5D3 Fab heavy chain variable domain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.843633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGPG LVKPSQSLSL TCTVTGFSIT SDYAWNWIRQ FPGKKLEWMG YINFDGGTTY NPSLRGRISI TRDTSKNQFF LQLRSVTPE DTATYYCATF YGAKGTLDYW GQGTSVTVSS AKTTPPSVYP LAPVCGDTSG SSVTLGCLVK GYFPEPVTLT W NSGSLSSG ...String: QVQLQESGPG LVKPSQSLSL TCTVTGFSIT SDYAWNWIRQ FPGKKLEWMG YINFDGGTTY NPSLRGRISI TRDTSKNQFF LQLRSVTPE DTATYYCATF YGAKGTLDYW GQGTSVTVSS AKTTPPSVYP LAPVCGDTSG SSVTLGCLVK GYFPEPVTLT W NSGSLSSG VHTFPAVLQS DLYTLSSSVT VTSSTWPSQS ITCNVAHPAS STKVDKKIEP RGP |
-Macromolecule #4: FAB heavy chain constant domain
| Macromolecule | Name: FAB heavy chain constant domain / type: protein_or_peptide / ID: 4 Details: The Fab domains were not resolved in the original structure and, therefore, were not modeled. With the implementation of an improved method, we can now resolve the domains within the CryoEM ...Details: The Fab domains were not resolved in the original structure and, therefore, were not modeled. With the implementation of an improved method, we can now resolve the domains within the CryoEM density map. However, given the absence of FAB information in the original structure, we opted to fit a representative FAB structure, utilizing PDB ID 7FAB as a reference. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.98684 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVQLEQSGPG LVRPSQTLSL TCTVSGTSFD DYYWTWVRQP PGRGLEWIGY VFYTGTTLLD PSLRGRVTML VNTSKNQFSL RLSSVTAAD TAVYYCARNL IAGGIDVWGQ GSLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN S GALTSGVH ...String: AVQLEQSGPG LVRPSQTLSL TCTVSGTSFD DYYWTWVRQP PGRGLEWIGY VFYTGTTLLD PSLRGRVTML VNTSKNQFSL RLSSVTAAD TAVYYCARNL IAGGIDVWGQ GSLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY FPEPVTVSWN S GALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEP |
-Macromolecule #5: Fab light chain constant domain
| Macromolecule | Name: Fab light chain constant domain / type: protein_or_peptide / ID: 5 Details: The Fab domains were not resolved in the original structure and, therefore, were not modeled. With the implementation of an improved method, we can now resolve the domains within the CryoEM ...Details: The Fab domains were not resolved in the original structure and, therefore, were not modeled. With the implementation of an improved method, we can now resolve the domains within the CryoEM density map. However, given the absence of FAB information in the original structure, we opted to fit a representative FAB structure, utilizing PDB ID 7FAB as a reference. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.866355 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QMTQSPSSLS ASVGDRVTIT CRASQSVSSA VAWYQQKPGK APKLLIYSAS SLYSGVPSRF SGSRSGTDYT LTISSLQPED FATYYCQQD GWSLITFGQG TKVEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP REAKVQWKVD NALQSGNSQE S VTEQDSKD ...String: QMTQSPSSLS ASVGDRVTIT CRASQSVSSA VAWYQQKPGK APKLLIYSAS SLYSGVPSRF SGSRSGTDYT LTISSLQPED FATYYCQQD GWSLITFGQG TKVEIKRTVA APSVFIFPPS DEQLKSGTAS VVCLLNNFYP REAKVQWKVD NALQSGNSQE S VTEQDSKD STYSLSSTLT LSKADYEKHK VYACEVTHQG LSSPVTKSFN RG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)