[English] 日本語
 Yorodumi
Yorodumi- EMDB-41840: Gaussian Mixture Models based single particle refinement - GPCR (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gaussian Mixture Models based single particle refinement - GPCR (Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 from EMPIAR-10786) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsubstance P receptor binding / substance P receptor activity / positive regulation of corticosterone secretion / tachykinin receptor activity / positive regulation of flagellated sperm motility / aggressive behavior / Tachykinin receptors bind tachykinins / sperm ejaculation / positive regulation of uterine smooth muscle contraction / insemination ...substance P receptor binding / substance P receptor activity / positive regulation of corticosterone secretion / tachykinin receptor activity / positive regulation of flagellated sperm motility / aggressive behavior / Tachykinin receptors bind tachykinins / sperm ejaculation / positive regulation of uterine smooth muscle contraction / insemination / positive regulation of synaptic transmission, cholinergic / detection of abiotic stimulus / operant conditioning / positive regulation of lymphocyte proliferation / tachykinin receptor signaling pathway / response to ozone / sperm head / positive regulation of action potential / response to auditory stimulus / smooth muscle contraction involved in micturition / positive regulation of acute inflammatory response / positive regulation of blood pressure / regulation of smooth muscle cell proliferation / positive regulation of vascular permeability / positive regulation of hormone secretion / regulation of smooth muscle cell migration / positive regulation of ossification / positive regulation of leukocyte migration / eating behavior / negative regulation of heart rate / response to pain / positive regulation of vasoconstriction / behavioral response to pain / angiotensin-mediated drinking behavior / associative learning / positive regulation of epithelial cell migration / response to electrical stimulus / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / long-term memory / D1 dopamine receptor binding / neuropeptide signaling pathway / intracellular transport / neuronal dense core vesicle / sperm flagellum / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / positive regulation of stress fiber assembly / response to hormone / sperm midpiece / sensory perception of pain / regulation of insulin secretion / cellular response to glucagon stimulus / response to progesterone / adenylate cyclase activator activity / trans-Golgi network membrane / positive regulation of epithelial cell proliferation / positive regulation of synaptic transmission, GABAergic / response to nicotine / negative regulation of inflammatory response to antigenic stimulus / cellular response to nerve growth factor stimulus / bone development / regulation of blood pressure / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / response to estradiol / adenylate cyclase-activating dopamine receptor signaling pathway Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
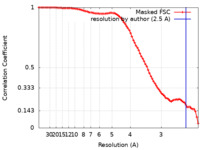
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Chen M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2024 Journal: Nat Methods / Year: 2024Title: Improving resolution and resolvability of single-particle cryoEM structures using Gaussian mixture models. Authors: Muyuan Chen / Michael F Schmid / Wah Chiu /  Abstract: Cryogenic electron microscopy is widely used in structural biology, but its resolution is often limited by the dynamics of the macromolecule. Here we developed a refinement protocol based on Gaussian ...Cryogenic electron microscopy is widely used in structural biology, but its resolution is often limited by the dynamics of the macromolecule. Here we developed a refinement protocol based on Gaussian mixture models that integrates particle orientation and conformation estimation and improves the alignment for flexible domains of protein structures. We demonstrated this protocol on multiple datasets, resulting in improved resolution and resolvability, locally and globally, by visual and quantitative measures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41840.map.gz emd_41840.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41840-v30.xml emd-41840-v30.xml emd-41840.xml emd-41840.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41840_fsc.xml emd_41840_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41840.png emd_41840.png | 117.5 KB | ||
| Filedesc metadata |  emd-41840.cif.gz emd-41840.cif.gz | 6.3 KB | ||
| Others |  emd_41840_half_map_1.map.gz emd_41840_half_map_1.map.gz emd_41840_half_map_2.map.gz emd_41840_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41840 http://ftp.pdbj.org/pub/emdb/structures/EMD-41840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41840 | HTTPS FTP |
-Validation report
| Summary document |  emd_41840_validation.pdf.gz emd_41840_validation.pdf.gz | 697.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41840_full_validation.pdf.gz emd_41840_full_validation.pdf.gz | 697.1 KB | Display | |
| Data in XML |  emd_41840_validation.xml.gz emd_41840_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_41840_validation.cif.gz emd_41840_validation.cif.gz | 20.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41840 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41840 | HTTPS FTP |
-Related structure data
| Related structure data |  8u26MC  8u28C  8u2cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41840.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41840.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.146 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41840_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41840_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Substance P bound to active human neurokinin 1 receptor in comple...
| Entire | Name: Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 |
|---|---|
| Components |
|
-Supramolecule #1: Substance P bound to active human neurokinin 1 receptor in comple...
| Supramolecule | Name: Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Reprocessing of EMPIAR-10786. Sample information is the same as EMD-24570, PDB-7RMH. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 141 KDa |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.907684 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NSKTEDQRNE EKAQREANKK IEKQLQKDKQ VYRATHRLLL LGADNSGKST IVKQMRILHG GSGGSGGTSG IFETKFQVDK VNFHMFDVG GQRDERRKWI QCFNDVTAII FVVDSSDYNR LQEALNLFKS IWNNRWLRTI SVILFLNKQD LLAEKVLAGK S KIEDYFPE ...String: NSKTEDQRNE EKAQREANKK IEKQLQKDKQ VYRATHRLLL LGADNSGKST IVKQMRILHG GSGGSGGTSG IFETKFQVDK VNFHMFDVG GQRDERRKWI QCFNDVTAII FVVDSSDYNR LQEALNLFKS IWNNRWLRTI SVILFLNKQD LLAEKVLAGK S KIEDYFPE FARYTTPEDA TPEPGEDPRV TRAKYFIRDE FLRISTASGD GRHYCYPHFT CAVDTENARR IFNDCRDIIQ RM HLRQYEL L UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.786566 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHLEV LFQGPEDQVD PRLIDGKGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIY AMHWGTDSRL LVSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR E GNVRVSRE ...String: MHHHHHHLEV LFQGPEDQVD PRLIDGKGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIY AMHWGTDSRL LVSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR E GNVRVSRE LAGHTGYLSC CRFLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KL WDVREGM CRQTFTGHES DINAICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYD DFNCNV WDALKADRAG VLAGHDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.398067 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSG SEDQVDPRLI DGK |
-Macromolecule #5: Substance-P receptor
| Macromolecule | Name: Substance-P receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.542867 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDASI DMDNVLPVDS DLSPNISTNT SEPNQFVQPA WQIVLWAAAY TVIVVTSVVG NVVVMWIILA HKRMRTVTNY FLVNLAFAE ASMAAFNTVV NFTYAVHNEW YYGLFYCKFH NFFPIAAVFA SIYSMTAVAF DRYMAIIHPL QPRLSATATK V VICVIWVL ...String: DYKDDDDASI DMDNVLPVDS DLSPNISTNT SEPNQFVQPA WQIVLWAAAY TVIVVTSVVG NVVVMWIILA HKRMRTVTNY FLVNLAFAE ASMAAFNTVV NFTYAVHNEW YYGLFYCKFH NFFPIAAVFA SIYSMTAVAF DRYMAIIHPL QPRLSATATK V VICVIWVL ALLLAFPQGY YSTTETMPSR VVCMIEWPEH PNKIYEKVYH ICVTVLIYFL PLLVIGYAYT VVGITLWASE IP GDSSDRY HEQVSAKRKV VKMMIVVVCT FAICWLPFHI FFLLPYINPD LYLKKFIQQV YLAIMWLAMS STMYNPIIYC CLN DRFRLG FKHAFRCCPF ISAGDYEGLE MKSTRYLQTQ GSVYKVSRLE TTISTVVGAH EEEPEDGPKA TPSSLDLTSN CSSR SDSKT MTESFSFSSN VLS UniProtKB: Substance-P receptor |
-Macromolecule #6: Protachykinin-1
| Macromolecule | Name: Protachykinin-1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.350629 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RPKPQQFFGL M UniProtKB: Protachykinin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8u26: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)