+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4181 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA Polymerase III initially transcribing complex | |||||||||
 Map data Map data | ITC map sharpened with relion_postprocess | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription / RNA Polymerase III / Pre-Initiation Complex / TFIIIB / Brf1 / Bdp1 / Enzyme | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase III core binding / TFIIA-class transcription factor complex binding / DNA-templated transcription open complex formation / RNA polymerase III transcription regulatory region sequence-specific DNA binding / RNA polymerase III preinitiation complex assembly / transcription factor TFIIIB complex / TFIIIC-class transcription factor complex binding / RNA polymerase I general transcription initiation factor binding / RNA polymerase III type 3 promoter sequence-specific DNA binding / regulation of transcription by RNA polymerase III ...RNA polymerase III core binding / TFIIA-class transcription factor complex binding / DNA-templated transcription open complex formation / RNA polymerase III transcription regulatory region sequence-specific DNA binding / RNA polymerase III preinitiation complex assembly / transcription factor TFIIIB complex / TFIIIC-class transcription factor complex binding / RNA polymerase I general transcription initiation factor binding / RNA polymerase III type 3 promoter sequence-specific DNA binding / regulation of transcription by RNA polymerase III / RNA polymerase III general transcription initiation factor activity / transcription factor TFIIA complex / RNA polymerase I preinitiation complex assembly / RNA Polymerase I Transcription Initiation / transcription preinitiation complex / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / DNA binding, bending / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA polymerase II general transcription initiation factor activity / termination of RNA polymerase III transcription / RNA Polymerase II Pre-transcription Events / transcription factor TFIID complex / RNA-templated transcription / Formation of TC-NER Pre-Incision Complex / RNA Polymerase I Promoter Escape / transcription initiation at RNA polymerase III promoter / termination of RNA polymerase I transcription / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / transcription initiation at RNA polymerase I promoter / Estrogen-dependent gene expression / transcription by RNA polymerase III / Dual incision in TC-NER / RNA polymerase I complex / RNA polymerase III complex / transcription elongation by RNA polymerase I / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / RNA polymerase II, core complex / tRNA transcription by RNA polymerase III / transcription by RNA polymerase I / nucleotidyltransferase activity / TBP-class protein binding / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / disordered domain specific binding / DNA-directed RNA polymerase activity / peroxisome / single-stranded DNA binding / ribosome biogenesis / transcription regulator complex / DNA-binding transcription factor binding / nucleic acid binding / transcription by RNA polymerase II / RNA polymerase II-specific DNA-binding transcription factor binding / protein dimerization activity / nucleotide binding / negative regulation of DNA-templated transcription / chromatin binding / regulation of DNA-templated transcription / nucleolus / positive regulation of transcription by RNA polymerase II / protein-containing complex / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
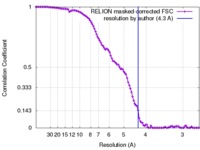
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Vorlaender MK / Khatter H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Molecular mechanism of promoter opening by RNA polymerase III. Authors: Matthias K Vorländer / Heena Khatter / Rene Wetzel / Wim J H Hagen / Christoph W Müller /  Abstract: RNA polymerase III (Pol III) and transcription factor IIIB (TFIIIB) assemble together on different promoter types to initiate the transcription of small, structured RNAs. Here we present structures ...RNA polymerase III (Pol III) and transcription factor IIIB (TFIIIB) assemble together on different promoter types to initiate the transcription of small, structured RNAs. Here we present structures of Pol III preinitiation complexes, comprising the 17-subunit Pol III and the heterotrimeric transcription factor TFIIIB, bound to a natural promoter in different functional states. Electron cryo-microscopy reconstructions, varying from 3.7 Å to 5.5 Å resolution, include two early intermediates in which the DNA duplex is closed, an open DNA complex, and an initially transcribing complex with RNA in the active site. Our structures reveal an extremely tight, multivalent interaction between TFIIIB and promoter DNA, and explain how TFIIIB recruits Pol III. Together, TFIIIB and Pol III subunit C37 activate the intrinsic transcription factor-like activity of the Pol III-specific heterotrimer to initiate the melting of double-stranded DNA, in a mechanism similar to that of the Pol II system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4181.map.gz emd_4181.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4181-v30.xml emd-4181-v30.xml emd-4181.xml emd-4181.xml | 47.9 KB 47.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4181_fsc.xml emd_4181_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4181.png emd_4181.png | 58.7 KB | ||
| Masks |  emd_4181_msk_1.map emd_4181_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4181.cif.gz emd-4181.cif.gz | 12.1 KB | ||
| Others |  emd_4181_additional.map.gz emd_4181_additional.map.gz | 2.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4181 http://ftp.pdbj.org/pub/emdb/structures/EMD-4181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4181 | HTTPS FTP |
-Related structure data
| Related structure data |  6f41MC  4180C  4182C  4183C  4184C  6f40C  6f42C  6f44C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10190 (Title: RNA Polymerase III initially transcribing complex / Data size: 1.6 TB / Data #1: Movie frames [micrographs - multiframe] EMPIAR-10190 (Title: RNA Polymerase III initially transcribing complex / Data size: 1.6 TB / Data #1: Movie frames [micrographs - multiframe]Data #2: Aligned sums of the movie frames without doseweighting [micrographs - single frame] Data #3: Doseweighted aligned sums of the movie frames [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ITC map sharpened with relion_postprocess | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4181_msk_1.map emd_4181_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: ITC map sharpened with LocScale
| File | emd_4181_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ITC map sharpened with LocScale | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : RNA Polymerase III initially transcribing complex
+Supramolecule #1: RNA Polymerase III initially transcribing complex
+Supramolecule #2: RNA Polymerase III
+Supramolecule #3: Transcription factors
+Supramolecule #4: Nucleic acids
+Macromolecule #1: DNA-directed RNA polymerase III subunit RPC1
+Macromolecule #2: DNA-directed RNA polymerase III subunit RPC2
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase III subunit RPC9
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase III subunit RPC8
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase III subunit RPC10
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: DNA-directed RNA polymerase III subunit RPC5
+Macromolecule #14: DNA-directed RNA polymerase III subunit RPC4
+Macromolecule #15: DNA-directed RNA polymerase III subunit RPC3
+Macromolecule #16: DNA-directed RNA polymerase III subunit RPC6
+Macromolecule #17: DNA-directed RNA polymerase III subunit RPC7
+Macromolecule #18: TATA-box-binding protein
+Macromolecule #19: Transcription factor IIIB 70 kDa subunit
+Macromolecule #20: Transcription factor TFIIIB component B''
+Macromolecule #21: RNA
+Macromolecule #22: Non-Template DNA
+Macromolecule #23: Template-DNA
+Macromolecule #24: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 61.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 142 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6f41: |
 Movie
Movie Controller
Controller





















 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)