[English] 日本語
 Yorodumi
Yorodumi- EMDB-41307: KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 cro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 crosslinked with its elongation ACP partner | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | polyketide synthase / antibody / BIOSYNTHETIC PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information6-deoxyerythronolide-B synthase / erythronolide synthase activity / macrolide biosynthetic process / fatty acid synthase activity / phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process / oxidoreductase activity Similarity search - Function | |||||||||||||||
| Biological species |  Saccharopolyspora erythraea (bacteria) Saccharopolyspora erythraea (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||||||||
 Authors Authors | Cogan DP / Soohoo AM / Chen M / Brodsky KL / Liu Y / Khosla C | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2025 Journal: Nat Chem Biol / Year: 2025Title: Structural basis for intermodular communication in assembly-line polyketide biosynthesis. Authors: Dillon P Cogan / Alexander M Soohoo / Muyuan Chen / Yan Liu / Krystal L Brodsky / Chaitan Khosla /  Abstract: Assembly-line polyketide synthases (PKSs) are modular multi-enzyme systems with considerable potential for genetic reprogramming. Understanding how they selectively transport biosynthetic ...Assembly-line polyketide synthases (PKSs) are modular multi-enzyme systems with considerable potential for genetic reprogramming. Understanding how they selectively transport biosynthetic intermediates along a defined sequence of active sites could be harnessed to rationally alter PKS product structures. To investigate functional interactions between PKS catalytic and substrate acyl carrier protein (ACP) domains, we employed a bifunctional reagent to crosslink transient domain-domain interfaces of a prototypical assembly line, the 6-deoxyerythronolide B synthase, and resolved their structures by single-particle cryogenic electron microscopy (cryo-EM). Together with statistical per-particle image analysis of cryo-EM data, we uncovered interactions between ketosynthase (KS) and ACP domains that discriminate between intra-modular and inter-modular communication while reinforcing the relevance of conformational asymmetry during the catalytic cycle. Our findings provide a foundation for the structure-based design of hybrid PKSs comprising biosynthetic modules from different naturally occurring assembly lines. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41307.map.gz emd_41307.map.gz | 171.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41307-v30.xml emd-41307-v30.xml emd-41307.xml emd-41307.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41307.png emd_41307.png | 27.7 KB | ||
| Filedesc metadata |  emd-41307.cif.gz emd-41307.cif.gz | 7 KB | ||
| Others |  emd_41307_half_map_1.map.gz emd_41307_half_map_1.map.gz emd_41307_half_map_2.map.gz emd_41307_half_map_2.map.gz | 318.2 MB 318.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41307 http://ftp.pdbj.org/pub/emdb/structures/EMD-41307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41307 | HTTPS FTP |
-Related structure data
| Related structure data |  8tjpMC  8tjnC  8tjoC  8tkoC  8tpwC  8tpxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41307.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41307.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.946 Å | ||||||||||||||||||||||||||||||||||||
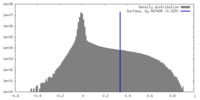
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41307_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
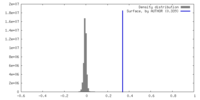
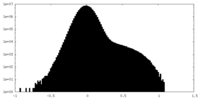
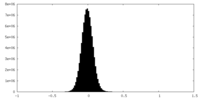
| Density Histograms |
-Half map: #1
| File | emd_41307_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
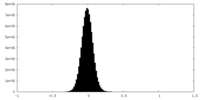
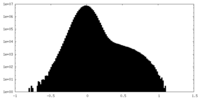
| Density Histograms |
- Sample components
Sample components
-Entire : KS-AT core of DEBS Module 3 crosslinked with its elongation ACP p...
| Entire | Name: KS-AT core of DEBS Module 3 crosslinked with its elongation ACP partner |
|---|---|
| Components |
|
-Supramolecule #1: KS-AT core of DEBS Module 3 crosslinked with its elongation ACP p...
| Supramolecule | Name: KS-AT core of DEBS Module 3 crosslinked with its elongation ACP partner type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Saccharopolyspora erythraea (bacteria) Saccharopolyspora erythraea (bacteria) |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: EryAII
| Macromolecule | Name: EryAII / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Saccharopolyspora erythraea (bacteria) Saccharopolyspora erythraea (bacteria) |
| Molecular weight | Theoretical: 99.76575 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVTDSEKVAE YLRRATLDLR AARQRIRELE SDPIAIVSMA CRLPGGVNTP QRLWELLREG GETLSGFPTD RGWDLARLHH PDPDNPGTS YVDKGGFLDD AAGFDAEFFG VSPREAAAMD PQQRLLLETS WELVENAGID PHSLRGTATG VFLGVAKFGY G EDTAAAED ...String: MVTDSEKVAE YLRRATLDLR AARQRIRELE SDPIAIVSMA CRLPGGVNTP QRLWELLREG GETLSGFPTD RGWDLARLHH PDPDNPGTS YVDKGGFLDD AAGFDAEFFG VSPREAAAMD PQQRLLLETS WELVENAGID PHSLRGTATG VFLGVAKFGY G EDTAAAED VEGYSVTGVA PAVASGRISY TMGLEGPSIS VDTACSSSLV ALHLAVESLR KGESSMAVVG GAAVMATPGV FV DFSRQRA LAADGRSKAF GAGADGFGFS EGVTLVLLER LSEARRNGHE VLAVVRGSAL NQDGASNGLS APSGPAQRRV IRQ ALESCG LEPGDVDAVE AHGTGTALGD PIEANALLDT YGRDRDADRP LWLGSVKSNI GHTQAAAGVT GLLKVVLALR NGEL PATLH VEEPTPHVDW SSGGVALLAG NQPWRRGERT RRAAVSAFGI SGTNAHVIVE EAPEREHRET TAHDGRPVPL VVSAR STAA LRAQAAQIAE LLERPDADLA GVGLGLATTR ARHEHRAAVV ASTREEAVRG LREIAAGAAT ADAVVEGVTE VDGRNV VFL FPGQGSQWAG MGAELLSSSP VFAGKIRACD ESMAPMQDWK VSDVLRQAPG APGLDRVDVV QPVLFAVMVS LAELWRS YG VEPAAVVGHS QGEIAAAHVA GALTLEDAAK LVVGRSRLMR SLSGEGGMAA VALGEAAVRE RLRPWQDRLS VAAVNGPR S VVVSGEPGAL RAFSEDCAAE GIRVRDIDVD YASHSPQIER VREELLETTG DIAPRPARVT FHSTVESRSM DGTELDARY WYRNLRETVR FADAVTRLAE SGYDAFIEVS PHPVVVQAVE EAVEEADGAE DAVVVGSLHR DGGDLSAFLR SMATAHVSGV DIRWDVALP GAAPFALPTY PFQRKRYWLQ PAAPAAASDE LAYRSSSVDK LAAALEHHHH HH UniProtKB: 6-deoxyerythronolide-B synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 11 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 30 s glow, 10 s hold, 15 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 4601 / Average exposure time: 6.45 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)