[English] 日本語
 Yorodumi
Yorodumi- EMDB-41306: Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 1 in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 1 in complex with antibody fragment 1B2: Crosslinked Intra-State 1 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | polyketide synthase / antibody / BIOSYNTHETIC PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Biological species |  Saccharopolyspora erythraea (bacteria) / Saccharopolyspora erythraea (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||||||||
 Authors Authors | Cogan DP / Soohoo AM / Chen M / Brodsky KL / Liu Y / Khosla C | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2025 Journal: Nat Chem Biol / Year: 2025Title: Structural basis for intermodular communication in assembly-line polyketide biosynthesis. Authors: Dillon P Cogan / Alexander M Soohoo / Muyuan Chen / Yan Liu / Krystal L Brodsky / Chaitan Khosla /  Abstract: Assembly-line polyketide synthases (PKSs) are modular multi-enzyme systems with considerable potential for genetic reprogramming. Understanding how they selectively transport biosynthetic ...Assembly-line polyketide synthases (PKSs) are modular multi-enzyme systems with considerable potential for genetic reprogramming. Understanding how they selectively transport biosynthetic intermediates along a defined sequence of active sites could be harnessed to rationally alter PKS product structures. To investigate functional interactions between PKS catalytic and substrate acyl carrier protein (ACP) domains, we employed a bifunctional reagent to crosslink transient domain-domain interfaces of a prototypical assembly line, the 6-deoxyerythronolide B synthase, and resolved their structures by single-particle cryogenic electron microscopy (cryo-EM). Together with statistical per-particle image analysis of cryo-EM data, we uncovered interactions between ketosynthase (KS) and ACP domains that discriminate between intra-modular and inter-modular communication while reinforcing the relevance of conformational asymmetry during the catalytic cycle. Our findings provide a foundation for the structure-based design of hybrid PKSs comprising biosynthetic modules from different naturally occurring assembly lines. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41306.map.gz emd_41306.map.gz | 256.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41306-v30.xml emd-41306-v30.xml emd-41306.xml emd-41306.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41306.png emd_41306.png | 31 KB | ||
| Filedesc metadata |  emd-41306.cif.gz emd-41306.cif.gz | 8.3 KB | ||
| Others |  emd_41306_half_map_1.map.gz emd_41306_half_map_1.map.gz emd_41306_half_map_2.map.gz emd_41306_half_map_2.map.gz | 475 MB 475 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41306 http://ftp.pdbj.org/pub/emdb/structures/EMD-41306 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41306 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41306 | HTTPS FTP |
-Related structure data
| Related structure data |  8tjoMC  8tjnC  8tjpC  8tkoC  8tpwC  8tpxC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41306.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41306.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
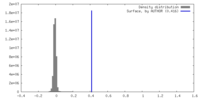
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41306_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
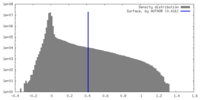
| Density Histograms |
-Half map: #1
| File | emd_41306_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
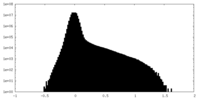
| Density Histograms |
- Sample components
Sample components
-Entire : Crosslinked DEBS Module 1 in complex with Antibody Fragment 1B2
| Entire | Name: Crosslinked DEBS Module 1 in complex with Antibody Fragment 1B2 |
|---|---|
| Components |
|
-Supramolecule #1: Crosslinked DEBS Module 1 in complex with Antibody Fragment 1B2
| Supramolecule | Name: Crosslinked DEBS Module 1 in complex with Antibody Fragment 1B2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Saccharopolyspora erythraea (bacteria) Saccharopolyspora erythraea (bacteria) |
| Molecular weight | Theoretical: 480 KDa |
-Macromolecule #1: EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6
| Macromolecule | Name: EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: 6-deoxyerythronolide-B synthase |
|---|---|
| Source (natural) | Organism:  Saccharopolyspora erythraea (bacteria) Saccharopolyspora erythraea (bacteria) |
| Molecular weight | Theoretical: 188.891219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASTDSEKVA EYLRRATLDL RAARQRIREL EGEPVAVVAM ACRLPGGVST PEEFWELLSE GRDAVAGLPT DRGWDLDSLF HPDPTRSGT AHQRGGGFLT EATAFDPAFF GMSPREALAV DPQQRLMLEL SWEVLERAGI PPTSLQASPT GVFVGLIPQE Y GPRLAEGG ...String: MASTDSEKVA EYLRRATLDL RAARQRIREL EGEPVAVVAM ACRLPGGVST PEEFWELLSE GRDAVAGLPT DRGWDLDSLF HPDPTRSGT AHQRGGGFLT EATAFDPAFF GMSPREALAV DPQQRLMLEL SWEVLERAGI PPTSLQASPT GVFVGLIPQE Y GPRLAEGG EGVEGYLMTG TTTSVASGRI AYTLGLEGPA ISVDTACSSS LVAVHLACQS LRRGESSLAM AGGVTVMPTP GM LVDFSRM NSLAPDGRCK AFSAGANGFG MAEGAGMLLL ERLSDARRNG HPVLAVLRGT AVNSDGASNG LSAPNGRAQV RVI QQALAE SGLGPADIDA VEAHGTGTRL GDPIEARALF EAYGRDREQP LHLGSVKSNL GHTQAAAGVA GVIKMVLAMR AGTL PRTLH ASERSKEIDW SSGAISLLDE PEPWPAGARP RRAGVSSFGI SGTNAHAIIE EAPQVVEGER VEAGDVVAPW VLSAS SAEG LRAQAARLAA HLREHPGQDP RDIAYSLATG RAALPHRAAF APVDESAALR VLDGLATGNA DGAAVGTSRA QQRAVF VFP GQGWQWAGMA VDLLDTSPVF AAALRECADA LEPHLDFEVI PFLRAEAARR EQDAALSTER VDVVQPVMFA VMVSLAS MW RAHGVEPAAV IGHSQGEIAA ACVAGALSLD DAARVVALRS RVIATMPGNK GMASIAAPAG EVRARIGDRV EIAAVNGP R SVVVAGDSDE LDRLVASCTT ECIRAKRLAV DYASHSSHVE TIRDALHAEL GEDFHPLPGF VPFFSTVTGR WTQPDELDA GYWYRNLRRT VRFADAVRAL AEQGYRTFLE VSAHPILTAA IEEIGDGSGA DLSAIHSLRR GDGSLADFGE ALSRAFAAGV AVDWESVHL GTGARRVPLP TYPFQRERVW LEPKPVARRS TEVDEVSALR YRIEWRPTGA GEPARLDGTW LVAKYAGTAD E TSTAAREA LESAGARVRE LVVDARCGRD ELAERLRSVG EVAGVLSLLA VDEAEPEEAP LALASLADTL SLVQAMVSAE LG CPLWTVT ESAVATGPFE RVRNAAHGAL WGVGRVIALE NPAVWGGLVD VPAGSVAELA RHLAAVVSGG AGEDQLALRA DGV YGRRWV RAAAPATDDE WKPTGTVLVT GGTGGVGGQI ARWLARRGAP HLLLVSRSGP DADGAGELVA ELEALGARTT VAAC DVTDR ESVRELLGGI GDDVPLSAVF HAAATLDDGT VDTLTGERIE RASRAKVLGA RNLHELTREL DLTAFVLFSS FASAF GAPG LGGYAPGNAY LDGLAQQRRS DGLPATAVAW GTWAGSGMAE GPVADRFRRH GVIEMPPETA CRALQNALDR AEVCPI VID VRWDRFLLAY TAQRPTRLFD EIDDARRAAP QAAAEPRVGA LASLPAPERE KALFELVRSH AAAVLGHASA ERVPADQ AF AELGVD(4HH)LSA LELRNRLGAA TGVRLPTTTV FDHPDVRTLA AHLTSELGSG TPAREASSAL RDGYRQAGVS GRVR SYLDL LAGLSDFREH FDGSDGFSLD LVDMADGPGE VTVICCAGTA AISGPHEFTR LAGALRGIAP VRAVPQPGYE EGEPL PSSM AAVAAVQADA VIRTQGDKPF VVAGHSAGAL MAYALATELL DRGHPPRGVV LIDVYPPGHQ DAMNAWLEEL TATLFD RET VRMDDTRLTA LGAYDRLTGQ WRPRETGLPT LLVSAGEPMG PWPDDSWKPT WPFEHDTVAV PGDHFTMVQE HADAIAR HI DAWLGGGNSS SVDKLAAALE HHHHHH |
-Macromolecule #2: Antibody Fragment 1B2, Heavy Chain
| Macromolecule | Name: Antibody Fragment 1B2, Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.447611 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEVQLVQSG GGLVQPGRSL RLSCTASGFT FGDYAMSWVR QAPGKGLEWV GFIRSKAYGG TTEYAASVKG RFTISRDDSK SIAYLQMNS LKTEDTAVYY CTRGGTLFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: MAEVQLVQSG GGLVQPGRSL RLSCTASGFT FGDYAMSWVR QAPGKGLEWV GFIRSKAYGG TTEYAASVKG RFTISRDDSK SIAYLQMNS LKTEDTAVYY CTRGGTLFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCAALVP RGSAHHHHHH AA DYKDDDD KA |
-Macromolecule #3: Antibody Fragment 1B2, Light Chain
| Macromolecule | Name: Antibody Fragment 1B2, Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.715832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LFAIPLVVPF YSHSALDVVM TQSPLSLPVT PGEPASISCR SSQSLLHSNG YNYLDWYLQK PGQSPQLLIY LGSNRASGVP DRFSGSGSG TDFTLKISRV EAEDVGVYYC MQSLQTPRLT FGPGTKVDIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN N FYPRGAKV ...String: LFAIPLVVPF YSHSALDVVM TQSPLSLPVT PGEPASISCR SSQSLLHSNG YNYLDWYLQK PGQSPQLLIY LGSNRASGVP DRFSGSGSG TDFTLKISRV EAEDVGVYYC MQSLQTPRLT FGPGTKVDIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN N FYPRGAKV QWKVDNALQS GNSQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 11 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 9523 / Average exposure time: 3.76 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)