+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | H1-nucleosome (chromatosome) | |||||||||
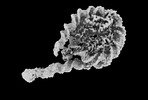
 Map data Map data | H1-nucleosome (chromatosome), 3DFlex reconstruction, locally filtered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / gene repression / epigenetics / chromatin / GENE REGULATION | |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Sauer PV / Pavlenko E / Nogales E / Poepsel S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Activation of automethylated PRC2 by dimerization on chromatin. Authors: Paul V Sauer / Egor Pavlenko / Trinity Cookis / Linda C Zirden / Juliane Renn / Ankush Singhal / Pascal Hunold / Michaela N Hoehne-Wiechmann / Olivia van Ray / Farnusch Kaschani / Markus ...Authors: Paul V Sauer / Egor Pavlenko / Trinity Cookis / Linda C Zirden / Juliane Renn / Ankush Singhal / Pascal Hunold / Michaela N Hoehne-Wiechmann / Olivia van Ray / Farnusch Kaschani / Markus Kaiser / Robert Hänsel-Hertsch / Karissa Y Sanbonmatsu / Eva Nogales / Simon Poepsel /   Abstract: Polycomb repressive complex 2 (PRC2) is an epigenetic regulator that trimethylates lysine 27 of histone 3 (H3K27me3) and is essential for embryonic development and cellular differentiation. H3K27me3 ...Polycomb repressive complex 2 (PRC2) is an epigenetic regulator that trimethylates lysine 27 of histone 3 (H3K27me3) and is essential for embryonic development and cellular differentiation. H3K27me3 is associated with transcriptionally repressed chromatin and is established when PRC2 is allosterically activated upon methyl-lysine binding by the regulatory subunit EED. Automethylation of the catalytic subunit enhancer of zeste homolog 2 (EZH2) stimulates its activity by an unknown mechanism. Here, we show that human PRC2 forms a dimer on chromatin in which an inactive, automethylated PRC2 protomer is the allosteric activator of a second PRC2 that is poised to methylate H3 of a substrate nucleosome. Functional assays support our model of allosteric trans-autoactivation via EED, suggesting a previously unknown mechanism mediating context-dependent activation of PRC2. Our work showcases the molecular mechanism of auto-modification-coupled dimerization in the regulation of chromatin-modifying complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41147.map.gz emd_41147.map.gz | 201.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41147-v30.xml emd-41147-v30.xml emd-41147.xml emd-41147.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41147.png emd_41147.png | 43 KB | ||
| Masks |  emd_41147_msk_1.map emd_41147_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41147.cif.gz emd-41147.cif.gz | 4.4 KB | ||
| Others |  emd_41147_additional_1.map.gz emd_41147_additional_1.map.gz emd_41147_additional_2.map.gz emd_41147_additional_2.map.gz emd_41147_additional_3.map.gz emd_41147_additional_3.map.gz emd_41147_half_map_1.map.gz emd_41147_half_map_1.map.gz emd_41147_half_map_2.map.gz emd_41147_half_map_2.map.gz | 109.1 MB 3 MB 3 MB 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41147 http://ftp.pdbj.org/pub/emdb/structures/EMD-41147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41147 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41147.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41147.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), 3DFlex reconstruction, locally filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41147_msk_1.map emd_41147_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
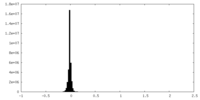
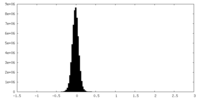
| Density Histograms |
-Additional map: H1-nucleosome (chromatosome), non-uniform refinement
| File | emd_41147_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
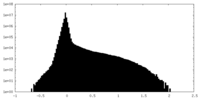
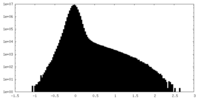
| Density Histograms |
-Additional map: H1-nucleosome (chromatosome), 3DFlex reconstruction, half map A
| File | emd_41147_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), 3DFlex reconstruction, half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
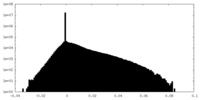
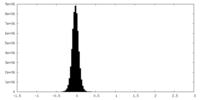
| Density Histograms |
-Additional map: H1-nucleosome (chromatosome), 3DFlex reconstruction, half map B
| File | emd_41147_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), 3DFlex reconstruction, half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
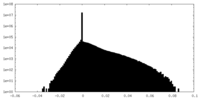
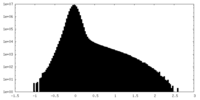
| Density Histograms |
-Half map: H1-nucleosome (chromatosome), non-uniform refinement, map half A
| File | emd_41147_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), non-uniform refinement, map half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: H1-nucleosome (chromatosome), non-uniform refinement, map half B
| File | emd_41147_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | H1-nucleosome (chromatosome), non-uniform refinement, map half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : H1-nucleosome (chromatosome)
| Entire | Name: H1-nucleosome (chromatosome) |
|---|---|
| Components |
|
-Supramolecule #1: H1-nucleosome (chromatosome)
| Supramolecule | Name: H1-nucleosome (chromatosome) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#13 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 290 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 44000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)