+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Automethylated PRC2 dimer bound to nucleosome | |||||||||
 Map data Map data | Automethylated PRC2 dimer bound to nucleosome 3DFlex reconstruction locally filtered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Histone methyl transferase / gene repression / epigenetics / chromatin / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationhepatocyte homeostasis / cellular response to trichostatin A / regulation of gliogenesis / negative regulation of striated muscle cell differentiation / regulation of kidney development / [histone H3]-lysine27 N-trimethyltransferase / response to tetrachloromethane / CAF-1 complex / histone H3K27 trimethyltransferase activity / negative regulation of keratinocyte differentiation ...hepatocyte homeostasis / cellular response to trichostatin A / regulation of gliogenesis / negative regulation of striated muscle cell differentiation / regulation of kidney development / [histone H3]-lysine27 N-trimethyltransferase / response to tetrachloromethane / CAF-1 complex / histone H3K27 trimethyltransferase activity / negative regulation of keratinocyte differentiation / negative regulation of retinoic acid receptor signaling pathway / cerebellar cortex development / primary miRNA binding / regulation of adaxial/abaxial pattern formation / random inactivation of X chromosome / skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration / histone H3K27 methyltransferase activity / sex chromatin / regulatory ncRNA-mediated heterochromatin formation / positive regulation of cell cycle G1/S phase transition / NURF complex / facultative heterochromatin formation / NuRD complex / regulation of cell fate specification / negative regulation of stem cell population maintenance / genomic imprinting / DNA replication-dependent chromatin assembly / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / ESC/E(Z) complex / negative regulation of stem cell differentiation / regulation of stem cell differentiation / protein-lysine N-methyltransferase activity / RSC-type complex / Polo-like kinase mediated events / cardiac muscle hypertrophy in response to stress / chromatin silencing complex / Transcription of E2F targets under negative control by DREAM complex / pronucleus / histone H3K9me2/3 reader activity / positive regulation of dendrite development / histone H3 methyltransferase activity / G1 to G0 transition / histone methyltransferase activity / : / ATPase complex / negative regulation of G1/S transition of mitotic cell cycle / DNA methylation-dependent constitutive heterochromatin formation / spinal cord development / negative regulation of gene expression, epigenetic / G1/S-Specific Transcription / synaptic transmission, GABAergic / lncRNA binding / histone deacetylase complex / Transcriptional Regulation by E2F6 / Sin3-type complex / positive regulation of MAP kinase activity / positive regulation of stem cell population maintenance / oligodendrocyte differentiation / RNA Polymerase I Transcription Initiation / negative regulation of transcription elongation by RNA polymerase II / G0 and Early G1 / positive regulation of protein serine/threonine kinase activity / positive regulation of GTPase activity / negative regulation of cell differentiation / positive regulation of epithelial to mesenchymal transition / ribonucleoprotein complex binding / subtelomeric heterochromatin formation / RNA polymerase II core promoter sequence-specific DNA binding / Cyclin E associated events during G1/S transition / pericentric heterochromatin / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / Cyclin A:Cdk2-associated events at S phase entry / nucleosome binding / keratinocyte differentiation / Regulation of TP53 Activity through Acetylation / heterochromatin / protein localization to chromatin / negative regulation of cytokine production involved in inflammatory response / Deposition of new CENPA-containing nucleosomes at the centromere / liver regeneration / B cell differentiation / negative regulation of cell migration / SUMOylation of chromatin organization proteins / Regulation of PTEN gene transcription / transcription corepressor binding / stem cell differentiation / cellular response to leukemia inhibitory factor / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / transcription coregulator activity / hippocampus development / Negative Regulation of CDH1 Gene Transcription / HDACs deacetylate histones / promoter-specific chromatin binding / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / negative regulation of transforming growth factor beta receptor signaling pathway / enzyme activator activity / G1/S transition of mitotic cell cycle Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Sauer PV / Pavlenko E / Nogales E / Poepsel S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Activation of automethylated PRC2 by dimerization on chromatin. Authors: Paul V Sauer / Egor Pavlenko / Trinity Cookis / Linda C Zirden / Juliane Renn / Ankush Singhal / Pascal Hunold / Michaela N Hoehne-Wiechmann / Olivia van Ray / Farnusch Kaschani / Markus ...Authors: Paul V Sauer / Egor Pavlenko / Trinity Cookis / Linda C Zirden / Juliane Renn / Ankush Singhal / Pascal Hunold / Michaela N Hoehne-Wiechmann / Olivia van Ray / Farnusch Kaschani / Markus Kaiser / Robert Hänsel-Hertsch / Karissa Y Sanbonmatsu / Eva Nogales / Simon Poepsel /   Abstract: Polycomb repressive complex 2 (PRC2) is an epigenetic regulator that trimethylates lysine 27 of histone 3 (H3K27me3) and is essential for embryonic development and cellular differentiation. H3K27me3 ...Polycomb repressive complex 2 (PRC2) is an epigenetic regulator that trimethylates lysine 27 of histone 3 (H3K27me3) and is essential for embryonic development and cellular differentiation. H3K27me3 is associated with transcriptionally repressed chromatin and is established when PRC2 is allosterically activated upon methyl-lysine binding by the regulatory subunit EED. Automethylation of the catalytic subunit enhancer of zeste homolog 2 (EZH2) stimulates its activity by an unknown mechanism. Here, we show that human PRC2 forms a dimer on chromatin in which an inactive, automethylated PRC2 protomer is the allosteric activator of a second PRC2 that is poised to methylate H3 of a substrate nucleosome. Functional assays support our model of allosteric trans-autoactivation via EED, suggesting a previously unknown mechanism mediating context-dependent activation of PRC2. Our work showcases the molecular mechanism of auto-modification-coupled dimerization in the regulation of chromatin-modifying complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41110.map.gz emd_41110.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41110-v30.xml emd-41110-v30.xml emd-41110.xml emd-41110.xml | 46.3 KB 46.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41110.png emd_41110.png | 41.5 KB | ||
| Masks |  emd_41110_msk_1.map emd_41110_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41110.cif.gz emd-41110.cif.gz | 9.5 KB | ||
| Others |  emd_41110_additional_1.map.gz emd_41110_additional_1.map.gz emd_41110_additional_2.map.gz emd_41110_additional_2.map.gz emd_41110_additional_3.map.gz emd_41110_additional_3.map.gz emd_41110_additional_4.map.gz emd_41110_additional_4.map.gz emd_41110_additional_5.map.gz emd_41110_additional_5.map.gz emd_41110_additional_6.map.gz emd_41110_additional_6.map.gz emd_41110_half_map_1.map.gz emd_41110_half_map_1.map.gz emd_41110_half_map_2.map.gz emd_41110_half_map_2.map.gz | 67.8 MB 1.2 MB 1.2 MB 1.2 MB 474.5 KB 1.1 MB 127.2 MB 127.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41110 http://ftp.pdbj.org/pub/emdb/structures/EMD-41110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41110 | HTTPS FTP |
-Related structure data
| Related structure data |  8t9gMC  8tasC  8tb9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41110.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41110.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Automethylated PRC2 dimer bound to nucleosome 3DFlex reconstruction locally filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.3 Å | ||||||||||||||||||||||||||||||||||||
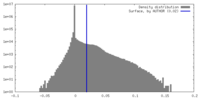
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41110_msk_1.map emd_41110_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Automethylated PRC2 dimer bound to nucleosome
+Supramolecule #1: Automethylated PRC2 dimer bound to nucleosome
+Supramolecule #2: Nucleosome
+Supramolecule #3: Proximal PRC2
+Supramolecule #4: Distal PRC2
+Macromolecule #1: Polycomb protein SUZ12
+Macromolecule #2: Histone-lysine N-methyltransferase EZH2
+Macromolecule #3: Polycomb protein EED
+Macromolecule #4: Histone-binding protein RBBP4
+Macromolecule #5: Zinc finger protein AEBP2
+Macromolecule #6: Histone H3.2
+Macromolecule #8: Histone H4
+Macromolecule #9: Histone H2A type 1
+Macromolecule #10: Histone H2B 1.1
+Macromolecule #12: activating methylated peptide
+Macromolecule #7: DNA (226-MER)
+Macromolecule #11: DNA (226-MER)
+Macromolecule #13: S-ADENOSYL-L-HOMOCYSTEINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Details: Streptavidin affinity grid |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 34000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8t9g: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
























 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)


