[English] 日本語
 Yorodumi
Yorodumi- EMDB-40986: Structure of a group II intron ribonucleoprotein in the pre-branc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | |||||||||
 Map data Map data | Overall refined map for pre-1f | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNP / RNA / Transferase-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  [Eubacterium] rectale (bacteria) [Eubacterium] rectale (bacteria) | |||||||||
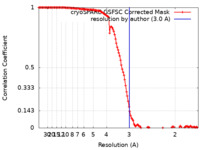
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Xu L / Liu T / Chung K / Pyle AM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural insights into intron catalysis and dynamics during splicing. Authors: Ling Xu / Tianshuo Liu / Kevin Chung / Anna Marie Pyle /  Abstract: The group II intron ribonucleoprotein is an archetypal splicing system with numerous mechanistic parallels to the spliceosome, including excision of lariat introns. Despite the importance of ...The group II intron ribonucleoprotein is an archetypal splicing system with numerous mechanistic parallels to the spliceosome, including excision of lariat introns. Despite the importance of branching in RNA metabolism, structural understanding of this process has remained elusive. Here we present a comprehensive analysis of three single-particle cryogenic electron microscopy structures captured along the splicing pathway. They reveal the network of molecular interactions that specifies the branchpoint adenosine and positions key functional groups to catalyse lariat formation and coordinate exon ligation. The structures also reveal conformational rearrangements of the branch helix and the mechanism of splice site exchange that facilitate the transition from branching to ligation. These findings shed light on the evolution of splicing and highlight the conservation of structural components, catalytic mechanism and dynamical strategies retained through time in premessenger RNA splicing machines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40986.map.gz emd_40986.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40986-v30.xml emd-40986-v30.xml emd-40986.xml emd-40986.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40986_fsc.xml emd_40986_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40986.png emd_40986.png | 58.2 KB | ||
| Masks |  emd_40986_msk_1.map emd_40986_msk_1.map emd_40986_msk_2.map emd_40986_msk_2.map | 216 MB 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40986.cif.gz emd-40986.cif.gz | 6.1 KB | ||
| Others |  emd_40986_additional_1.map.gz emd_40986_additional_1.map.gz emd_40986_additional_2.map.gz emd_40986_additional_2.map.gz emd_40986_half_map_1.map.gz emd_40986_half_map_1.map.gz emd_40986_half_map_2.map.gz emd_40986_half_map_2.map.gz | 203.9 MB 203.7 MB 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40986 http://ftp.pdbj.org/pub/emdb/structures/EMD-40986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40986 | HTTPS FTP |
-Related structure data
| Related structure data |  8t2sMC  8t2rC  8t2tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40986.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40986.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall refined map for pre-1f | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40986_msk_1.map emd_40986_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
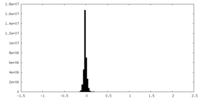
| Density Histograms |
-Mask #2
| File |  emd_40986_msk_2.map emd_40986_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
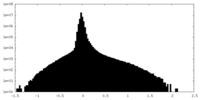
| Density Histograms |
-Additional map: Locally refined map (right) for pre-1f
| File | emd_40986_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined map (right) for pre-1f | ||||||||||||
| Projections & Slices |
| ||||||||||||
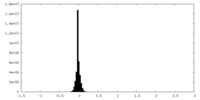
| Density Histograms |
-Additional map: Locally refined map (left) for pre-1f
| File | emd_40986_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined map (left) for pre-1f | ||||||||||||
| Projections & Slices |
| ||||||||||||
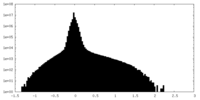
| Density Histograms |
-Half map: Overall map (half A)
| File | emd_40986_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall map (half_A) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Overall map (half B)
| File | emd_40986_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall map (half_B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Intron RNP complex in pre-1f
| Entire | Name: Intron RNP complex in pre-1f |
|---|---|
| Components |
|
-Supramolecule #1: Intron RNP complex in pre-1f
| Supramolecule | Name: Intron RNP complex in pre-1f / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  [Eubacterium] rectale (bacteria) [Eubacterium] rectale (bacteria) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Group II intron reverse transcriptase/maturase
| Macromolecule | Name: Group II intron reverse transcriptase/maturase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  [Eubacterium] rectale (bacteria) [Eubacterium] rectale (bacteria) |
| Molecular weight | Theoretical: 49.083914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDTSNLMEQI LSSDNLNRAY LQVVRNKGAE GVDGMKYTEL KEHLAKNGET IKGQLRTRKY KPQPARRVEI PKPDGGVRNL GVPTVTDRF IQQAIAQVLT PIYEEQFHDH SYGFRPNRCA QQAILTALNI MNDGNDWIVD IDLEKFFDTV NHDKLMTLIG R TIKDGDVI ...String: MDTSNLMEQI LSSDNLNRAY LQVVRNKGAE GVDGMKYTEL KEHLAKNGET IKGQLRTRKY KPQPARRVEI PKPDGGVRNL GVPTVTDRF IQQAIAQVLT PIYEEQFHDH SYGFRPNRCA QQAILTALNI MNDGNDWIVD IDLEKFFDTV NHDKLMTLIG R TIKDGDVI SIVRKYLVSG IMIDDEYEDS IVGTPQGGNL SPLLANIMLN ELDKEMEKRG LNFVRYADDC IIMVGSEMSA NR VMRNISR FIEEKLGLKV NMTKSKVDRP SGLKYLGFGF YFDPRAHQFK AKPHAKSVAK FKKRMKELTC RSWGVSNSYK VEK LNQLIR GWINYFKIGS MKTLCKELDS RIRYRLRMCI WKQWKTPQNQ EKNLVKLGID RNTARRVAYT GKRIAYVCNK GAVN VAISN KRLASFGLIS MLDYYIEKCV TC UniProtKB: Group II intron reverse transcriptase/maturase |
-Macromolecule #2: RNA (551-MER)
| Macromolecule | Name: RNA (551-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  [Eubacterium] rectale (bacteria) [Eubacterium] rectale (bacteria) |
| Molecular weight | Theoretical: 211.194406 KDa |
| Sequence | String: GAGCUCAUUU CUUUGUUUGC GCGCCAUGGG CGCGCUCUAA CGGGUGUAAG UCCCGAACAU GCCCAGGUAG UGGGAAAUGU AUAGCCGAA CAGCAAGGGU GUCUACUGUG AGGUGGAAUC UGAAGGAAGC UGUAAGCGAA UCUCUGGUCC GACGGACAGA A AUCGCAUA ...String: GAGCUCAUUU CUUUGUUUGC GCGCCAUGGG CGCGCUCUAA CGGGUGUAAG UCCCGAACAU GCCCAGGUAG UGGGAAAUGU AUAGCCGAA CAGCAAGGGU GUCUACUGUG AGGUGGAAUC UGAAGGAAGC UGUAAGCGAA UCUCUGGUCC GACGGACAGA A AUCGCAUA UAAGGCUAGG CUUCGAGUGA UAAGCUGGCA AAGAACAGUG AAGUCUAAUA ACUACCACGU UUGUAGAAGC AG AGUAAAU GCGGCGGAUA UAUGGAGAGA AAGAGCGUGC ACCUUAAGCG UGGAGGUCUC ACAGAGGUUU CAUUAGCCUA GUA ACAACG AACUGUGAGA AGUCAGCCGA GCCCAUAGUA GUGAAGAAGU CUCUGUAAUG GGGAUGGAGC GAAGGGGCGA ACAA UCAAU CAGUUUGAGA AUGUCUCGUA UUGCAGAAAU GACAACAUCU GCCGUAACCA AUCGGGUAAA AGGUGGUCAA AUCAA GCGA GACGGAAAGG AAAGAACGCA UGGACACAAG UAAUCUAAUU UCGGUUAGAU UACUACAUCG AAAAGUGUGU UACUUG UUA AAUUGAUUGA ACCGCCGUAU ACGGAACCGU ACGUACGGUG GUGUGAGAGG UCGGAAUUUC UCAAUUAAGA GAAAUUC UU CCUACUCGAU |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 11 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: AMMONIUM ION
| Macromolecule | Name: AMMONIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: NH4 |
|---|---|
| Molecular weight | Theoretical: 18.038 Da |
| Chemical component information |  ChemComp-NH4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)