+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3996 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | N4 ring of PilQ from Thermus thermophilus | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Thermus thermophilus HB27 (bacteria) Thermus thermophilus HB27 (bacteria) | |||||||||
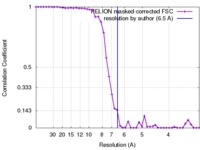
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | D'Imprima E / Vonck J / Sanchez R | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Cryo-EM structure of the bifunctional secretin complex of . Authors: Edoardo D'Imprima / Ralf Salzer / Ramachandra M Bhaskara / Ricardo Sánchez / Ilona Rose / Lennart Kirchner / Gerhard Hummer / Werner Kühlbrandt / Janet Vonck / Beate Averhoff /  Abstract: Secretins form multimeric channels across the outer membrane of Gram-negative bacteria that mediate the import or export of substrates and/or extrusion of type IV pili. The secretin complex of is an ...Secretins form multimeric channels across the outer membrane of Gram-negative bacteria that mediate the import or export of substrates and/or extrusion of type IV pili. The secretin complex of is an oligomer of the 757-residue PilQ protein, essential for DNA uptake and pilus extrusion. Here, we present the cryo-EM structure of this bifunctional complex at a resolution of ~7 Å using a new reconstruction protocol. Thirteen protomers form a large periplasmic domain of six stacked rings and a secretin domain in the outer membrane. A homology model of the PilQ protein was fitted into the cryo-EM map. A crown-like structure outside the outer membrane capping the secretin was found not to be part of PilQ. Mutations in the secretin domain disrupted the crown and abolished DNA uptake, suggesting a central role of the crown in natural transformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3996.map.gz emd_3996.map.gz | 510.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3996-v30.xml emd-3996-v30.xml emd-3996.xml emd-3996.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3996_fsc.xml emd_3996_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3996.png emd_3996.png | 14.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3996 http://ftp.pdbj.org/pub/emdb/structures/EMD-3996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3996 | HTTPS FTP |
-Validation report
| Summary document |  emd_3996_validation.pdf.gz emd_3996_validation.pdf.gz | 217.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3996_full_validation.pdf.gz emd_3996_full_validation.pdf.gz | 216.4 KB | Display | |
| Data in XML |  emd_3996_validation.xml.gz emd_3996_validation.xml.gz | 9.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3996 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3996 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3996.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3996.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.63 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
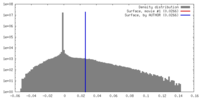
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PilQ complex with 13-fold symmetry
| Entire | Name: PilQ complex with 13-fold symmetry |
|---|---|
| Components |
|
-Supramolecule #1: PilQ complex with 13-fold symmetry
| Supramolecule | Name: PilQ complex with 13-fold symmetry / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB27 (bacteria) Thermus thermophilus HB27 (bacteria) |
| Molecular weight | Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: blotting for 8-10 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Lower energy threshold: 18 eV / Energy filter - Upper energy threshold: 18 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-45 / Average exposure time: 9.0 sec. / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 30675 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.2 mm / Nominal magnification: 20000 |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)