[English] 日本語
 Yorodumi
Yorodumi- EMDB-3907: The electron crystallography structure of the cAMP-bound potassiu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3907 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | |||||||||
 Map data Map data | PCO (Projective ConstraintOptimization) - refined 3D volume of MloK1 with cAMP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MloK1 / MlotiK1 / potassium channel / CNBD / cytoplasmic domains / PCO refinement / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellularly cyclic nucleotide-activated monoatomic cation channel activity / potassium channel activity / cAMP binding / protein-containing complex binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mesorhizobium loti (bacteria) / Mesorhizobium loti (bacteria) /  Mesorhizobium loti MAFF303099 (bacteria) Mesorhizobium loti MAFF303099 (bacteria) | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Kowal J / Biyani N | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer. Authors: Julia Kowal / Nikhil Biyani / Mohamed Chami / Sebastian Scherer / Andrzej J Rzepiela / Paul Baumgartner / Vikrant Upadhyay / Crina M Nimigean / Henning Stahlberg /   Abstract: Eukaryotic cyclic nucleotide-modulated channels perform their diverse physiological roles by opening and closing their pores to ions in response to cyclic nucleotide binding. We here present a ...Eukaryotic cyclic nucleotide-modulated channels perform their diverse physiological roles by opening and closing their pores to ions in response to cyclic nucleotide binding. We here present a structural model for the cyclic nucleotide-modulated potassium channel homolog from Mesorhizobium loti, MloK1, determined from 2D crystals in the presence of lipids. Even though crystals diffract electrons to only ∼10 Å, using cryoelectron microscopy (cryo-EM) and recently developed computational methods, we have determined a 3D map of full-length MloK1 in the presence of cyclic AMP (cAMP) at ∼4.5 Å isotropic 3D resolution. The structure provides a clear picture of the arrangement of the cyclic nucleotide-binding domains with respect to both the pore and the putative voltage sensor domains when cAMP is bound, and reveals a potential gating mechanism in the context of the lipid-embedded channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3907.map.gz emd_3907.map.gz | 8.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3907-v30.xml emd-3907-v30.xml emd-3907.xml emd-3907.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3907_1.png emd_3907_1.png emd_3907_2.png emd_3907_2.png | 207.5 KB 184.7 KB | ||
| Filedesc metadata |  emd-3907.cif.gz emd-3907.cif.gz | 7.2 KB | ||
| Others |  emd_3907_additional.map.gz emd_3907_additional.map.gz | 6.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3907 http://ftp.pdbj.org/pub/emdb/structures/EMD-3907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3907 | HTTPS FTP |
-Related structure data
| Related structure data |  6eo1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3907.map.gz / Format: CCP4 / Size: 9.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3907.map.gz / Format: CCP4 / Size: 9.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PCO (Projective ConstraintOptimization) - refined 3D volume of MloK1 with cAMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.955 Å / Y: 0.955 Å / Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
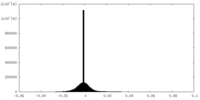
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Back-projected 3D volume of MloK1 with cAMP
| File | emd_3907_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Back-projected 3D volume of MloK1 with cAMP | ||||||||||||
| Projections & Slices |
| ||||||||||||
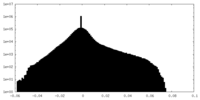
| Density Histograms |
- Sample components
Sample components
-Entire : MloK1 tetramer
| Entire | Name: MloK1 tetramer |
|---|---|
| Components |
|
-Supramolecule #1: MloK1 tetramer
| Supramolecule | Name: MloK1 tetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Cyclic nucleotide-modulated potassium channel in the presence of cAMP ligand, reconstituted into 2D lipid membrane crystals. |
|---|---|
| Source (natural) | Organism:  Mesorhizobium loti (bacteria) / Organ: Membrane Mesorhizobium loti (bacteria) / Organ: Membrane |
| Molecular weight | Theoretical: 148 KDa |
-Macromolecule #1: Cyclic nucleotide-gated potassium channel mll3241
| Macromolecule | Name: Cyclic nucleotide-gated potassium channel mll3241 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mesorhizobium loti MAFF303099 (bacteria) / Organ: Membrane Mesorhizobium loti MAFF303099 (bacteria) / Organ: Membrane |
| Molecular weight | Theoretical: 37.766297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVLPFLRIY APLNAVLAAP GLLAVAALTI PDMSGRSRLA LAALLAVIWG AYLLQLAATL LKRRAGVVRD RTPKIAIDVL AVLVPLAAF LLDGSPDWSL YCAVWLLKPL RDSTFFPVLG RVLANEARNL IGVTTLFGVV LFAVALAAYV IERDIQPEKF G SIPQAMWW ...String: MSVLPFLRIY APLNAVLAAP GLLAVAALTI PDMSGRSRLA LAALLAVIWG AYLLQLAATL LKRRAGVVRD RTPKIAIDVL AVLVPLAAF LLDGSPDWSL YCAVWLLKPL RDSTFFPVLG RVLANEARNL IGVTTLFGVV LFAVALAAYV IERDIQPEKF G SIPQAMWW AVVTLSTTGY GDTIPQSFAG RVLAGAVMMS GIGIFGLWAG ILATGFYQEV RRGDFVRNWQ LVAAVPLFQK LG PAVLVEI VRALRARTVP AGAVICRIGE PGDRMFFVVE GSVSVATPNP VELGPGAFFG EMALISGEPR SATVSAATTV SLL SLHSAD FQMLCSSSPE IAEIFRKTAL ERRGAAASA UniProtKB: Cyclic nucleotide-gated potassium channel mll3241 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 20 mM KCl, 20 mM Tris-HCl pH 7.6, 1 mM BaCl2, 1 mM EDTA, 0.2 mM cAMP |
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: 3.5 second-blotting. |
| Crystal formation | Lipid protein ratio: 0.8 / Lipid mixture: E.coli polar lipids / Instrument: dialysis buttons / Atmosphere: dialysis buffer / Temperature: 293.0 K / Time: 5.0 DAY Details: DM solubilized MloK1 sample was mixed with E. coli polar lipid extract (Avanti Polar Lipids) at a lipid to protein ratio of 0.8 and dialyzed against detergent free buffer. 2D crystals of the ...Details: DM solubilized MloK1 sample was mixed with E. coli polar lipid extract (Avanti Polar Lipids) at a lipid to protein ratio of 0.8 and dialyzed against detergent free buffer. 2D crystals of the lipid embedded protein were obtained within 5 days. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | pixel size 1.3 A/pix |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 30 / Number real images: 346 / Average exposure time: 16.0 sec. / Average electron dose: 45.0 e/Å2 Details: Each image was dose-fractionated in 40 frames (16 sec in total, 0.4-sec frames). The dose rate was set to ~5 counts/sec/physical-pixel (~2.8 e-/s/A2)leading to a total dose of ~45 e-/A2. Pixel size was 1.3A/pix. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.3 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 50000 / Camera length: 800 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN / Tilt angle: 0.0, 55.0 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1-355 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The initial model was obtained using Modeller (Sali and Blundell, 1993); in particular, the missing fragments were generated for the previously published PDB 4CHV model. This starting model was refined using the Rosetta for cryo-EM package (DiMaio et al., 2015). The symmetry of the channel was restrained during optimization runs (performed following the package tutorial (Wang and DiMaio,2015)). A model with a high fit score to the cryo-EM map and a low energy, as defined by the Rosetta force field, was selected from 100 Rosetta models generated and refined further. Several rounds of manual refinement with Coot (Emsley et al.,2010) and global optimization with Phenix (real_space_refine method (Afonine et al., 2013)) were carried out. Secondary structure constraints were imposed to stabilize the fold of helices and b-sheets during the global optimization. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: fit energy |
| Output model |  PDB-6eo1: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)