+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3887 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AcrB in a SMALP | |||||||||
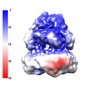
 Map data Map data | AcrB in SMA polymer. Map filtered by local resolution in RELION 2.1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Parmar M / Rawson S / Scarff CA / Goldman A / Dafforn TR / Muench SP / Postis VLG | |||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Biomembr / Year: 2018 Journal: Biochim Biophys Acta Biomembr / Year: 2018Title: Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Authors: Mayuriben Parmar / Shaun Rawson / Charlotte A Scarff / Adrian Goldman / Timothy R Dafforn / Stephen P Muench / Vincent L G Postis /   Abstract: The field of membrane protein structural biology has been revolutionized over the last few years with a number of high profile structures being solved using cryo-EM including Piezo, Ryanodine ...The field of membrane protein structural biology has been revolutionized over the last few years with a number of high profile structures being solved using cryo-EM including Piezo, Ryanodine receptor, TRPV1 and the Glutamate receptor. Further developments in the EM field hold the promise of even greater progress in terms of greater resolution, which for membrane proteins is still typically within the 4-7Å range. One advantage of a cryo-EM approach is the ability to study membrane proteins in more "native" like environments for example proteoliposomes, amphipols and nanodiscs. Recently, styrene maleic acid co-polymers (SMA) have been used to extract membrane proteins surrounded by native lipids (SMALPs) maintaining a more natural environment. We report here the structure of the Escherichia coli multidrug efflux transporter AcrB in a SMALP scaffold to sub-nm resolution, with the resulting map being consistent with high resolution crystal structures and other EM derived maps. However, both the C-terminal helix (TM12) and TM7 are poorly defined in the map. These helices are at the exterior of the helical bundle and form the greater interaction with the native lipids and SMA polymer and may represent a more dynamic region of the protein. This work shows the promise of using an SMA approach for single particle cryo-EM studies to provide sub-nm structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3887.map.gz emd_3887.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3887-v30.xml emd-3887-v30.xml emd-3887.xml emd-3887.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3887.png emd_3887.png | 76.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3887 http://ftp.pdbj.org/pub/emdb/structures/EMD-3887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3887 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3887.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3887.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AcrB in SMA polymer. Map filtered by local resolution in RELION 2.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Escherichia coli multidrug efflux transporter AcrB in a SMALP platform
| Entire | Name: Escherichia coli multidrug efflux transporter AcrB in a SMALP platform |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia coli multidrug efflux transporter AcrB in a SMALP platform
| Supramolecule | Name: Escherichia coli multidrug efflux transporter AcrB in a SMALP platform type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 340 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Low-pass filtered EM map |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 8.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 26480 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: OTHER / Details: RELION auto-refinement |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















