+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of human NuA4/TIP60 complex | |||||||||
 マップデータ マップデータ | human TIP60 complex overall half map1 | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Remodeler / Histone Acetyltransferase Complex / NuA4 / TIP60 / TRANSCRIPTION | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報piccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / sperm DNA condensation / histone chaperone activity / establishment of protein localization to chromatin / transcription factor TFTC complex / npBAF complex ...piccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / sperm DNA condensation / histone chaperone activity / establishment of protein localization to chromatin / transcription factor TFTC complex / npBAF complex / R2TP complex / brahma complex / dynein axonemal particle / neural retina development / GBAF complex / regulation of G0 to G1 transition / Swr1 complex / RPAP3/R2TP/prefoldin-like complex / protein antigen binding / chromatin-protein adaptor activity / RSC-type complex / Ino80 complex / blastocyst formation / regulation of nucleotide-excision repair / regulation of double-strand break repair / box C/D snoRNP assembly / SAGA complex / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / protein folding chaperone complex / positive regulation of T cell differentiation / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / positive regulation of double-strand break repair / spinal cord development / negative regulation of gene expression, epigenetic / DNA repair-dependent chromatin remodeling / NuA4 histone acetyltransferase complex / regulation of chromosome organization / positive regulation of stem cell population maintenance / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of RNA splicing / Transcriptional Regulation by E2F6 / Regulation of MITF-M-dependent genes involved in pigmentation / regulation of G1/S transition of mitotic cell cycle / TFIID-class transcription factor complex binding / regulation of DNA replication / MLL1 complex / regulation of embryonic development / Telomere Extension By Telomerase / negative regulation of cell differentiation / somatic stem cell population maintenance / spermatid development / enzyme-substrate adaptor activity / positive regulation of myoblast differentiation / positive regulation of double-strand break repair via homologous recombination / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / Deposition of new CENPA-containing nucleosomes at the centromere / telomere maintenance / DNA helicase activity / transcription initiation-coupled chromatin remodeling / positive regulation of DNA repair / TBP-class protein binding / replication fork / double-strand break repair via homologous recombination / DNA Damage Recognition in GG-NER / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / transcription coregulator activity / positive regulation of cell differentiation / cellular response to estradiol stimulus / chromatin DNA binding / Formation of the beta-catenin:TCF transactivating complex / helicase activity / euchromatin / negative regulation of canonical Wnt signaling pathway / ADP binding / beta-catenin binding / kinetochore / DNA Damage/Telomere Stress Induced Senescence / 加水分解酵素; 酸無水物に作用; 酸無水物に作用・細胞または細胞小器官の運動に関与 / positive regulation of protein import into nucleus / RMTs methylate histone arginines / nuclear matrix / UCH proteinases / cellular response to UV / positive regulation of canonical Wnt signaling pathway / transcription corepressor activity / unfolded protein binding / nucleosome / site of double-strand break / protein folding / nervous system development / HATs acetylate histones / chromatin organization / ATPase binding / response to ethanol / spermatogenesis / histone binding / nuclear membrane / regulation of apoptotic process 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
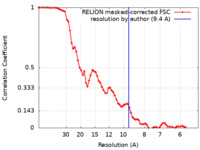
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 9.4 Å | |||||||||
 データ登録者 データ登録者 | Chen K / Wang L / Yu Z / Yu J / Ren Y / Wang Q / Xu Y | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2024 ジャーナル: Nat Commun / 年: 2024タイトル: Structure of the human TIP60 complex. 著者: Ke Chen / Li Wang / Zishuo Yu / Jiali Yu / Yulei Ren / Qianmin Wang / Yanhui Xu /  要旨: Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell ...Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell cycle control, and embryonic development. Here we report the cryo-electron microscopy structures of the human TIP60 complex with the core subcomplex and TRRAP module refined to 3.2-Å resolution. The structures show that EP400 acts as a backbone integrating the motor module, the ARP module, and the TRRAP module. The RUVBL1-RUVBL2 hexamer serves as a rigid core for the assembly of EP400 ATPase and YL1 in the motor module. In the ARP module, an ACTL6A-ACTB heterodimer and an extra ACTL6A make hydrophobic contacts with EP400 HSA helix, buttressed by network interactions among DMAP1, EPC1, and EP400. The ARP module stably associates with the motor module but is flexibly tethered to the TRRAP module, exhibiting a unique feature of human TIP60. The architecture of the nucleosome-bound human TIP60 reveals an unengaged nucleosome that is located between the core subcomplex and the TRRAP module. Our work illustrates the molecular architecture of human TIP60 and provides architectural insights into how this complex is bound by the nucleosome. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_38703.map.gz emd_38703.map.gz | 27.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-38703-v30.xml emd-38703-v30.xml emd-38703.xml emd-38703.xml | 32.5 KB 32.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_38703_fsc.xml emd_38703_fsc.xml | 7.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_38703.png emd_38703.png | 26.4 KB | ||
| Filedesc metadata |  emd-38703.cif.gz emd-38703.cif.gz | 12.5 KB | ||
| その他 |  emd_38703_half_map_1.map.gz emd_38703_half_map_1.map.gz emd_38703_half_map_2.map.gz emd_38703_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38703 http://ftp.pdbj.org/pub/emdb/structures/EMD-38703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38703 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_38703_validation.pdf.gz emd_38703_validation.pdf.gz | 785 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_38703_full_validation.pdf.gz emd_38703_full_validation.pdf.gz | 784.6 KB | 表示 | |
| XML形式データ |  emd_38703_validation.xml.gz emd_38703_validation.xml.gz | 12.8 KB | 表示 | |
| CIF形式データ |  emd_38703_validation.cif.gz emd_38703_validation.cif.gz | 17.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38703 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38703 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8xvgMC  8xvtC  8xvvC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_38703.map.gz / 形式: CCP4 / 大きさ: 30.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_38703.map.gz / 形式: CCP4 / 大きさ: 30.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | human TIP60 complex overall half map1 | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.668 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: human TIP60 complex overall half map1
| ファイル | emd_38703_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | human TIP60 complex overall half map1 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: human TIP60 complex overall half map2
| ファイル | emd_38703_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | human TIP60 complex overall half map2 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Structure of human NuA4/TIP60 complex
+超分子 #1: Structure of human NuA4/TIP60 complex
+分子 #1: Isoform 2 of Transformation/transcription domain-associated protein
+分子 #2: RuvB-like 1
+分子 #3: RuvB-like 2
+分子 #4: DNA methyltransferase 1-associated protein 1
+分子 #5: Isoform 2 of E1A-binding protein p400
+分子 #6: Actin-like protein 6A
+分子 #7: ACTB protein (Fragment)
+分子 #8: Isoform 2 of Enhancer of polycomb homolog 1
+分子 #9: Vacuolar protein sorting-associated protein 72 homolog
+分子 #10: INOSITOL HEXAKISPHOSPHATE
+分子 #11: ADENOSINE-5'-DIPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.8000000000000003 µm 最小 デフォーカス(公称値): 1.5 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)