+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The architecture of human NuA4/TIP60-nucleosome complex | |||||||||
 Map data Map data | overall map of human TIP60-NCP complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Remodeler / Histone Acetyltransferase Complex / NuA4 / TIP60 / TRANSCRIPTION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
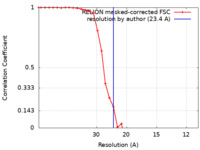
| Method | single particle reconstruction / cryo EM / Resolution: 23.4 Å | |||||||||
 Authors Authors | Chen K / Wang L / Yu Z / Yu J / Ren Y / Wang Q / Xu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of the human TIP60 complex. Authors: Ke Chen / Li Wang / Zishuo Yu / Jiali Yu / Yulei Ren / Qianmin Wang / Yanhui Xu /  Abstract: Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell ...Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell cycle control, and embryonic development. Here we report the cryo-electron microscopy structures of the human TIP60 complex with the core subcomplex and TRRAP module refined to 3.2-Å resolution. The structures show that EP400 acts as a backbone integrating the motor module, the ARP module, and the TRRAP module. The RUVBL1-RUVBL2 hexamer serves as a rigid core for the assembly of EP400 ATPase and YL1 in the motor module. In the ARP module, an ACTL6A-ACTB heterodimer and an extra ACTL6A make hydrophobic contacts with EP400 HSA helix, buttressed by network interactions among DMAP1, EPC1, and EP400. The ARP module stably associates with the motor module but is flexibly tethered to the TRRAP module, exhibiting a unique feature of human TIP60. The architecture of the nucleosome-bound human TIP60 reveals an unengaged nucleosome that is located between the core subcomplex and the TRRAP module. Our work illustrates the molecular architecture of human TIP60 and provides architectural insights into how this complex is bound by the nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60547.map.gz emd_60547.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60547-v30.xml emd-60547-v30.xml emd-60547.xml emd-60547.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60547_fsc.xml emd_60547_fsc.xml | 3 KB | Display |  FSC data file FSC data file |
| Images |  emd_60547.png emd_60547.png | 21.7 KB | ||
| Filedesc metadata |  emd-60547.cif.gz emd-60547.cif.gz | 3.9 KB | ||
| Others |  emd_60547_additional_1.map.gz emd_60547_additional_1.map.gz emd_60547_additional_2.map.gz emd_60547_additional_2.map.gz emd_60547_additional_3.map.gz emd_60547_additional_3.map.gz emd_60547_half_map_1.map.gz emd_60547_half_map_1.map.gz emd_60547_half_map_2.map.gz emd_60547_half_map_2.map.gz | 792.4 KB 707.2 KB 756.4 KB 1.4 MB 1.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60547 http://ftp.pdbj.org/pub/emdb/structures/EMD-60547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60547 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60547.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60547.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | overall map of human TIP60-NCP complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.56 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: The core subcomplex of human TIP60-NCP complex
| File | emd_60547_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The core subcomplex of human TIP60-NCP complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: The NCP-containing module of human TIP60-NCP complex
| File | emd_60547_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The NCP-containing module of human TIP60-NCP complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: The TRRAP module of human TIP60-NCP complex
| File | emd_60547_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The TRRAP module of human TIP60-NCP complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: human TIP60-NCP complex overall half map2
| File | emd_60547_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human TIP60-NCP complex overall half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the architecture of human TIP60-nucleosome complex
| Entire | Name: the architecture of human TIP60-nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: the architecture of human TIP60-nucleosome complex
| Supramolecule | Name: the architecture of human TIP60-nucleosome complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)