+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The TRRAP module of human NuA4/TIP60 complex | |||||||||
 Map data Map data | The TRRAP module of human TIP60 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Remodeler / Histone Acetyltransferase Complex / NuA4 / TIP60 / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscription factor TFTC complex / Swr1 complex / protein antigen binding / regulation of double-strand break repair / SAGA complex / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / DNA repair-dependent chromatin remodeling / regulation of RNA splicing / NuA4 histone acetyltransferase complex / regulation of DNA repair ...transcription factor TFTC complex / Swr1 complex / protein antigen binding / regulation of double-strand break repair / SAGA complex / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / DNA repair-dependent chromatin remodeling / regulation of RNA splicing / NuA4 histone acetyltransferase complex / regulation of DNA repair / positive regulation of double-strand break repair via homologous recombination / transcription coregulator activity / Formation of the beta-catenin:TCF transactivating complex / helicase activity / DNA Damage/Telomere Stress Induced Senescence / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / nucleosome / HATs acetylate histones / chromatin organization / regulation of apoptotic process / regulation of cell cycle / Ub-specific processing proteases / nuclear speck / hydrolase activity / DNA repair / chromatin binding / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / Golgi apparatus / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
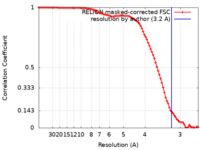
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Chen K / Wang L / Yu Z / Yu J / Ren Y / Wang Q / Xu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of the human TIP60 complex. Authors: Ke Chen / Li Wang / Zishuo Yu / Jiali Yu / Yulei Ren / Qianmin Wang / Yanhui Xu /  Abstract: Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell ...Mammalian TIP60 is a multi-functional enzyme with histone acetylation and histone dimer exchange activities. It plays roles in diverse cellular processes including transcription, DNA repair, cell cycle control, and embryonic development. Here we report the cryo-electron microscopy structures of the human TIP60 complex with the core subcomplex and TRRAP module refined to 3.2-Å resolution. The structures show that EP400 acts as a backbone integrating the motor module, the ARP module, and the TRRAP module. The RUVBL1-RUVBL2 hexamer serves as a rigid core for the assembly of EP400 ATPase and YL1 in the motor module. In the ARP module, an ACTL6A-ACTB heterodimer and an extra ACTL6A make hydrophobic contacts with EP400 HSA helix, buttressed by network interactions among DMAP1, EPC1, and EP400. The ARP module stably associates with the motor module but is flexibly tethered to the TRRAP module, exhibiting a unique feature of human TIP60. The architecture of the nucleosome-bound human TIP60 reveals an unengaged nucleosome that is located between the core subcomplex and the TRRAP module. Our work illustrates the molecular architecture of human TIP60 and provides architectural insights into how this complex is bound by the nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38720.map.gz emd_38720.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38720-v30.xml emd-38720-v30.xml emd-38720.xml emd-38720.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38720_fsc.xml emd_38720_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_38720.png emd_38720.png | 31.1 KB | ||
| Filedesc metadata |  emd-38720.cif.gz emd-38720.cif.gz | 10.1 KB | ||
| Others |  emd_38720_half_map_1.map.gz emd_38720_half_map_1.map.gz emd_38720_half_map_2.map.gz emd_38720_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38720 http://ftp.pdbj.org/pub/emdb/structures/EMD-38720 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38720 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38720 | HTTPS FTP |
-Related structure data
| Related structure data |  8xvvMC  8xvgC  8xvtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38720.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38720.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The TRRAP module of human TIP60 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.334 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map2 of the TRRAP module of human TIP60 complex
| File | emd_38720_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 of the TRRAP module of human TIP60 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1 of the TRRAP module of human TIP60 complex
| File | emd_38720_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1 of the TRRAP module of human TIP60 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The TRRAP module of human NuA4/TIP60 complex
| Entire | Name: The TRRAP module of human NuA4/TIP60 complex |
|---|---|
| Components |
|
-Supramolecule #1: The TRRAP module of human NuA4/TIP60 complex
| Supramolecule | Name: The TRRAP module of human NuA4/TIP60 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform 2 of E1A-binding protein p400
| Macromolecule | Name: Isoform 2 of E1A-binding protein p400 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 340.198062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQI TLAPLPLPSP TSPGFQFSAQ PRRFEHGSPS YIQVTSPLSQ QVQTQSPTQP SPGPGQALQN VRAGAPGPGL G LCSSSPTG ...String: MHHGTGPQNV QHQLQRSRAC PGSEGEEQPA HPNPPPSPAA PFAPSASPSA PQSPSYQIQQ LMNRSPATGQ NVNITLQSVG PVVGGNQQI TLAPLPLPSP TSPGFQFSAQ PRRFEHGSPS YIQVTSPLSQ QVQTQSPTQP SPGPGQALQN VRAGAPGPGL G LCSSSPTG GFVDASVLVR QISLSPSSGG HFVFQDGSGL TQIAQGAQVQ LQHPGTPITV RERRPSQPHT QSGGTIHHLG PQ SPAAAGG AGLQPLASPS HITTANLPPQ ISSIIQGQLV QQQQVLQGPP LPRPLGFERT PGVLLPGAGG AAGFGMTSPP PPT SPSRTA VPPGLSSLPL TSVGNTGMKK VPKKLEEIPP ASPEMAQMRK QCLDYHYQEM QALKEVFKEY LIELFFLQHF QGNM MDFLA FKKKHYAPLQ AYLRQNDLDI EEEEEEEEEE EEKSEVINDE QQALAGSLVA GAGSTVETDL FKRQQAMPST GMAEQ SKRP RLEVGHQGVV FQHPGADAGV PLQQLMPTAQ GGMPPTPQAA QLAGQRQSQQ QYDPSTGPPV QNAASLHTPL PQLPGR LPP AGVPTAALSS ALQFAQQPQV VEAQTQLQIP VKTQQPNVPI PAPPSSQLPI PPSQPAQLAL HVPTPGKVQV QASQLSS LP QMVASTRLPV DPAPPCPRPL PTSSTSSLAP VSGSGPGPSP ARSSPVNRPS SATNKALSPV TSRTPGVVAS APTKPQSP A QNATSSQDSS QDTLTEQITL ENQVHQRIAE LRKAGLWSQR RLPKLQEAPR PKSHWDYLLE EMQWMATDFA QERRWKVAA AKKLVRTVVR HHEEKQLREE RGKKEEQSRL RRIAASTARE IECFWSNIEQ VVEIKLRVEL EEKRKKALNL QKVSRRGKEL RPKGFDALQ ESSLDSGMSG RKRKASISLT DDEVDDEEET IEEEEANEGV VDHQTELSNL AKEAELPLLD LMKLYEGAFL P SSQWPRPK PDGEDTSGEE DADDCPGDRE SRKDLVLIDS LFIMDQFKAA ERMNIGKPNA KDIADVTAVA EAILPKGSAR VT TSVKFNA PSLLYGALRD YQKIGLDWLA KLYRKNLNGI LADEAGLGKT VQIIAFFAHL ACNEGNWGPH LVVVRSCNIL KWE LELKRW CPGLKILSYI GSHRELKAKR QEWAEPNSFH VCITSYTQFF RGLTAFTRVR WKCLVIDEMQ RVKGMTERHW EAVF TLQSQ QRLLLIDSPL HNTFLELWTM VHFLVPGISR PYLSSPLRAP SEESQDYYHK VVIRLHRVTQ PFILRRTKRD VEKQL TKKY EHVLKCRLSN RQKALYEDVI LQPGTQEALK SGHFVNVLSI LVRLQRICNH PGLVEPRHPG SSYVAGPLEY PSASLI LKA LERDFWKEAD LSMFDLIGLE NKITRHEAEL LSKKKIPRKL MEEISTSAAP AARPAAAKLK ASRLFQPVQY GQKPEGR TV AFPSTHPPRT AAPTTASAAP QGPLRGRPPI ATFSANPEAK AAAAPFQTSQ ASASAPRHQP ASASSTAASP AHPAKLRA Q TTAQASTPGQ PPPQPQAPSH AAGQSALPQR LVLPSQAQAR LPSGEVVKIA QLASITGPQS RVAQPETPVT LQFQGSKFT LSHSQLRQLT AGQPLQLQGS VLQIVSAPGQ PYLRAPGPVV MQTVSQAGAV HGALGSKPPA GGPSPAPLTP QVGVPGRVAV NALAVGEPG TASKPASPIG GPTQEEKTRL LKERLDQIYL VNERRCSQAP VYGRDLLRIC ALPSHGRVQW RGSLDGRRGK E AGPAHSYT SSSESPSELM LTLCRCGESL QDVIDRVAFV IPPVVAAPPS LRVPRPPPLY SHRMRILRQG LREHAAPYFQ QL RQTTAPR LLQFPELRLV QFDSGKLEAL AILLQKLKSE GRRVLILSQM ILMLDILEMF LNFHYLTYVR IDENASSEQR QEL MRSFNR DRRIFCAILS THSRTTGINL VEADTVVFYD NDLNPVMDAK AQEWCDRIGR CKDIHIYRLV SGNSIEEKLL KNGT KDLIR EVAAQGNDYS MAFLTQRTIQ ELFEVYSPMD DAGFPVKAEE FVVLSQEPSV TETIAPKIAR PFIEALKSIE YLEED AQKS AQEGVLGPHT DALSSDSENM PCDEEPSQLE ELADFMEQLT PIEKYALNYL ELFHTSIEQE KERNSEDAVM TAVRAW EFW NLKTLQEREA RLRLEQEEAE LLTYTREDAY SMEYVYEDVD GQTEVMPLWT PPTPPQDDSD IYLDSVMCLM YEATPIP EA KLPPVYVRKE RKRHKTDPSA AGRKKKQRHG EAVVPPRSLF DRATPGLLKI RREGKEQKKN ILLKQQVPFA KPLPTFAK P TAEPGQDNPE WLISEDWALL QAVKQLLELP LNLTIVSPAH TPNWDLVSDV VNSCSRIYRS SKQCRNRYEN VIIPREEGK SKNNRPLRTS QIYAQDENAT HTQLYTSHFD LMKMTAGKRS PPIKPLLGMN PFQKNPKHAS VLAESGINYD KPLPPIQVAS LRAERIAKE KKALADQQKA QQPAVAQPPP PQPQPPPPPQ QPPPPLPQPQ AAGSQPPAGP PAVQPQPQPQ PQTQPQPVQA P AKAQPAIT TGGSAAVLAG TIKTSVTGTS MPTGAVSGNV IVNTIAGVPA ATFQSINKRL ASPVAPGALT TPGGSAPAQV VH TQPPPRA VGSPATATPD LVSMATTQGV RAVTSVTASA VVTTNLTPVQ TPARSLVPQV SQATGVQLPG KTITPAHFQL LRQ QQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQTTTTSQVQ VPQIQGQAQS PAQIKAVGKL TPEHLIKMQK QKLQMPPQPP PPQA QSAPP QPTAQVQVQT SQPPQQQSPQ LTTVTAPRPG ALLTGTTVAN LQVARLTRVP TSQLQAQGQM QTQAPQPAQV ALAKP PVVS VPAAVVSSPG VTTLPMNVAG ISVAIGQPQK AAGQTVVAQP VHMQQLLKLK QQAVQQQKAI QPQAAQGPAA VQQKIT AQQ ITTPGAQQKV AYAAQPALKT QFLTTPISQA QKLAGAQQVQ TQIQVAKLPQ VVQQQTPVAS IQQVASASQQ ASPQTVA LT QATAAGQQVQ MIPAVTATAQ VVQQKLIQQQ VVTTASAPLQ TPGAPNPAQV PASSDSPSQQ PKLQMRVPAV RLKTPTKP P CQ UniProtKB: E1A-binding protein p400 |
-Macromolecule #2: Isoform 2 of Transformation/transcription domain-associated protein
| Macromolecule | Name: Isoform 2 of Transformation/transcription domain-associated protein type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 434.949906 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFVATQGAT VVDQTTLMKK YLQFVAALTD VNTPDETKLK MMQEVSENFE NVTSSPQYST FLEHIIPRFL TFLQDGEVQF LQEKPAQQL RKLVLEIIHR IPTNEHLRPH TKNVLSVMFR FLETENEENV LICLRIIIEL HKQFRPPITQ EIHHFLDFVK Q IYKELPKV ...String: MAFVATQGAT VVDQTTLMKK YLQFVAALTD VNTPDETKLK MMQEVSENFE NVTSSPQYST FLEHIIPRFL TFLQDGEVQF LQEKPAQQL RKLVLEIIHR IPTNEHLRPH TKNVLSVMFR FLETENEENV LICLRIIIEL HKQFRPPITQ EIHHFLDFVK Q IYKELPKV VNRYFENPQV IPENTVPPPE MVGMITTIAV KVNPEREDSE TRTHSIIPRG SLSLKVLAEL PIIVVLMYQL YK LNIHNVV AEFVPLIMNT IAIQVSAQAR QHKLYNKELY ADFIAAQIKT LSFLAYIIRI YQELVTKYSQ QMVKGMLQLL SNC PAETAH LRKELLIAAK HILTTELRNQ FIPCMDKLFD ESILIGSGYT ARETLRPLAY STLADLVHHV RQHLPLSDLS LAVQ LFAKN IDDESLPSSI QTMSCKLLLN LVDCIRSKSE QESGNGRDVL MRMLEVFVLK FHTIARYQLS AIFKKCKPQS ELGAV EAAL PGVPTAPAAP GPAPSPAPVP APPPPPPPPP PATPVTPAPV PPFEKQGEKD KEDKQTFQVT DCRSLVKTLV CGVKTI TWG ITSCKAPGEA QFIPNKQLQP KETQIYIKLV KYAMQALDIY QVQIAGNGQT YIRVANCQTV RMKEEKEVLE HFAGVFT MM NPLTFKEIFQ TTVPYMVERI SKNYALQIVA NSFLANPTTS ALFATILVEY LLDRLPEMGS NVELSNLYLK LFKLVFGS V SLFAAENEQM LKPHLHKIVN SSMELAQTAK EPYNYFLLLR ALFRSIGGGS HDLLYQEFLP LLPNLLQGLN MLQSGLHKQ HMKDLFVELC LTVPVRLSSL LPYLPMLMDP LVSALNGSQT LVSQGLRTLE LCVDNLQPDF LYDHIQPVRA ELMQALWRTL RNPADSISH VAYRVLGKFG GSNRKMLKES QKLHYVVTEV QGPSITVEFS DCKASLQLPM EKAIETALDC LKSANTEPYY R RQAWEVIK CFLVAMMSLE DNKHALYQLL AHPNFTEKTI PNVIISHRYK AQDTPARKTF EQALTGAFMS AVIKDLRPSA LP FVASLIR HYTMVAVAQQ CGPFLLPCYQ VGSQPSTAMF HSEENGSKGM DPLVLIDAIA ICMAYEEKEL CKIGEVALAV IFD VASIIL GSKERACQLP LFSYIVERLC ACCYEQAWYA KLGGVVSIKF LMERLPLTWV LQNQQTFLKA LLFVMMDLTG EVSN GAVAM AKTTLEQLLM RCATPLKDEE RAEEIVAAQE KSFHHVTHDL VREVTSPNST VRKQAMHSLQ VLAQVTGKSV TVIME PHKE VLQDMVPPKK HLLRHQPANA QIGLMEGNTF CTTLQPRLFT MDLNVVEHKV FYTELLNLCE AEDSALTKLP CYKSLP SLV PLRIAALNAL AACNYLPQSR EKIIAALFKA LNSTNSELQE AGEACMRKFL EGATIEVDQI HTHMRPLLMM LGDYRSL TL NVVNRLTSVT RLFPNSFNDK FCDQMMQHLR KWMEVVVITH KGGQRSDGNE MKICSAIINL FHLIPAAPQT LVKPLLEV V MKTERAMLIE AGSPFREPLI KFLTRHPSQT VELFMMEATL NDPQWSRMFM SFLKHKDARP LRDVLAANPN RFITLLLPG GAQTAVRPGS PSTSTMRLDL QFQAIKIISI IVKNDDSWLA SQHSLVSQLR RVWVSENFQE RHRKENMAAT NWKEPKLLAY CLLNYCKRN YGDIELLFQL LRAFTGRFLC NMTFLKEYME EEIPKNYSIA QKRALFFRFV DFNDPNFGDE LKAKVLQHIL N PAFLYSFE KGEGEQLLGP PNPEGDNPES ITSVFITKVL DPEKQADMLD SLRIYLLQYA TLLVEHAPHH IHDNNKNRNS KL RRLMTFA WPCLLSKACV DPACKYSGHL LLAHIIAKFA IHKKIVLQVF HSLLKAHAME ARAIVRQAMA ILTPAVPARM EDG HQMLTH WTRKIIVEEG HTVPQLVHIL HLIVQHFKVY YPVRHHLVQH MVSAMQRLGF TPSVTIEQRR LAVDLSEVVI KWEL QRIKD QQPDSDMDPN SSGEGVNSVS SSIKRGLSVD SAQEVKRFRT ATGAISAVFG RSQSLPGADS LLAKPIDKQH TDTVV NFLI RVACQVNDNT NTAGSPGEVL SRRCVNLLKT ALRPDMWPKS ELKLQWFDKL LMTVEQPNQV NYGNICTGLE VLSFLL TVL QSPAILSSFK PLQRGIAACM TCGNTKVLRA VHSLLSRLMS IFPTEPSTSS VASKYEELEC LYAAVGKVIY EGLTNYE KA TNANPSQLFG TLMILKSACS NNPSYIDRLI SVFMRSLQKM VREHLNPQAA SGSTEATSGT SELVMLSLEL VKTRLAVM S MEMRKNFIQA ILTSLIEKSP DAKILRAVVK IVEEWVKNNS PMAANQTPTL REKSILLVKM MTYIEKRFPE DLELNAQFL DLVNYVYRDE TLSGSELTAK LEPAFLSGLR CAQPLIRAKF FEVFDNSMKR RVYERLLYVT CSQNWEAMGN HFWIKQCIEL LLAVCEKST PIGTSCQGAM LPSITNVINL ADSHDRAAFA MVTHVKQEPR ERENSESKEE DVEIDIELAP GDQTSTPKTK E LSEKDIGN QLHMLTNRHD KFLDTLREVK TGALLSAFVQ LCHISTTLAE KTWVQLFPRL WKILSDRQQH ALAGEISPFL CS GSHQVQR DCQPSALNCF VEAMSQCVPP IPIRPCVLKY LGKTHNLWFR STLMLEHQAF EKGLSLQIKP KQTTEFYEQE SIT PPQQEI LDSLAELYSL LQEEDMWAGL WQKRCKYSET ATAIAYEQHG FFEQAQESYE KAMDKAKKEH ERSNASPAIF PEYQ LWEDH WIRCSKELNQ WEALTEYGQS KGHINPYLVL ECAWRVSNWT AMKEALVQVE VSCPKEMAWK VNMYRGYLAI CHPEE QQLS FIERLVEMAS SLAIREWRRL PHVVSHVHTP LLQAAQQIIE LQEAAQINAG LQPTNLGRNN SLHDMKTVVK TWRNRL PIV SDDLSHWSSI FMWRQHHYQA IVTAYENSSQ HDPSSNNAML GVHASASAII QYGKIARKQG LVNVALDILS RIHTIPT VP IVDCFQKIRQ QVKCYLQLAG VMGKNECMQG LEVIESTNLK YFTKEMTAEF YALKGMFLAQ INKSEEANKA FSAAVQMH D VLVKAWAMWG DYLENIFVKE RQLHLGVSAI TCYLHACRHQ NESKSRKYLA KVLWLLSFDD DKNTLADAVD KYCIGVPPI QWLAWIPQLL TCLVGSEGKL LLNLISQVGR VYPQAVYFPI RTLYLTLKIE QRERYKSDPG PIRATAPMWR CSRIMHMQRE LHPTLLSSL EGIVDQMVWF RENWHEEVLR QLQQGLAKCY SVAFEKSGAV SDAKITPHTL NFVKKLVSTF GVGLENVSNV S TMFSSAAS ESLARRAQAT AQDPVFQKLK GQFTTDFDFS VPGSMKLHNL ISKLKKWIKI LEAKTKQLPK FFLIEEKCRF LS NFSAQTA EVEIPGEFLM PKPTHYYIKI ARFMPRVEIV QKHNTAARRL YIRGHNGKIY PYLVMNDACL TESRREERVL QLL RLLNPC LEKRKETTKR HLFFTVPRVV AVSPQMRLVE DNPSSLSLVE IYKQRCAKKG IEHDNPISRY YDRLATVQAR GTQA SHQVL RDILKEVQSN MVPRSMLKEW ALHTFPNATD YWTFRKMFTI QLALIGFAEF VLHLNRLNPE MLQIAQDTGK LNVAY FRFD INDATGDLDA NRPVPFRLTP NISEFLTTIG VSGPLTASMI AVARCFAQPN FKVDGILKTV LRDEIIAWHK KTQEDT SSP LSAAGQPENM DSQQLVSLVQ KAVTAIMTRL HNLAQFEGGE SKVNTLVAAA NSLDNLCRMD PAWHPWL UniProtKB: Transformation/transcription domain-associated protein |
-Macromolecule #3: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)