+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3867 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sacbrood virus of honeybee - expansion state II | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | empty particle / icosahedral virus particle / iflavirus / expanded particle / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane / viral capsid / RNA helicase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / structural molecule activity / proteolysis / RNA binding ...host cell membrane / viral capsid / RNA helicase activity / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / structural molecule activity / proteolysis / RNA binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Sacbrood virus Sacbrood virus | |||||||||
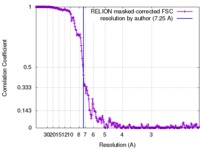
| Method | single particle reconstruction / cryo EM / Resolution: 7.25 Å | |||||||||
 Authors Authors | Plevka P / Prochazkova M | |||||||||
| Funding support |  Czech Republic, 2 items Czech Republic, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Virion structure and genome delivery mechanism of sacbrood honeybee virus. Authors: Michaela Procházková / Tibor Füzik / Karel Škubník / Jana Moravcová / Zorica Ubiparip / Antonín Přidal / Pavel Plevka /  Abstract: Infection by sacbrood virus (SBV) from the family Iflaviridae is lethal to honey bee larvae but only rarely causes the collapse of honey bee colonies. Despite the negative effect of SBV on honey ...Infection by sacbrood virus (SBV) from the family Iflaviridae is lethal to honey bee larvae but only rarely causes the collapse of honey bee colonies. Despite the negative effect of SBV on honey bees, the structure of its particles and mechanism of its genome delivery are unknown. Here we present the crystal structure of SBV virion and show that it contains 60 copies of a minor capsid protein (MiCP) attached to the virion surface. No similar MiCPs have been previously reported in any of the related viruses from the order Picornavirales. The location of the MiCP coding sequence within the SBV genome indicates that the MiCP evolved from a C-terminal extension of a major capsid protein by the introduction of a cleavage site for a virus protease. The exposure of SBV to acidic pH, which the virus likely encounters during cell entry, induces the formation of pores at threefold and fivefold axes of the capsid that are 7 Å and 12 Å in diameter, respectively. This is in contrast to vertebrate picornaviruses, in which the pores along twofold icosahedral symmetry axes are currently considered the most likely sites for genome release. SBV virions lack VP4 subunits that facilitate the genome delivery of many related dicistroviruses and picornaviruses. MiCP subunits induce liposome disruption in vitro, indicating that they are functional analogs of VP4 subunits and enable the virus genome to escape across the endosome membrane into the cell cytoplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3867.map.gz emd_3867.map.gz | 262.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3867-v30.xml emd-3867-v30.xml emd-3867.xml emd-3867.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3867_fsc.xml emd_3867_fsc.xml | 18.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_3867.png emd_3867.png | 284 KB | ||
| Filedesc metadata |  emd-3867.cif.gz emd-3867.cif.gz | 6.2 KB | ||
| Others |  emd_3867_additional.map.gz emd_3867_additional.map.gz | 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3867 http://ftp.pdbj.org/pub/emdb/structures/EMD-3867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3867 | HTTPS FTP |
-Related structure data
| Related structure data |  6eh1MC  3863C  3865C  3866C  3881C  5lsfC  5oypC  6egvC  6egxC  6eiwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3867.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3867.map.gz / Format: CCP4 / Size: 620.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 3867 additional.map
| File | emd_3867_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sacbrood virus
| Entire | Name:  Sacbrood virus Sacbrood virus |
|---|---|
| Components |
|
-Supramolecule #1: Sacbrood virus
| Supramolecule | Name: Sacbrood virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Isolated from honeybee pupae / NCBI-ID: 89463 / Sci species name: Sacbrood virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 278.86 Å / T number (triangulation number): 3 |
-Macromolecule #1: structural protein VP1
| Macromolecule | Name: structural protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sacbrood virus Sacbrood virus |
| Molecular weight | Theoretical: 24.27948 KDa |
| Sequence | String: IKDILRRPVL LFNHVELDPD YTGFFIPIMP PSRMMQYKSG DKETSFQRLI GRTPQAAIMN LFRFWRGSLR YTIIIHSTDG HPIYVTHVP HTGNRVYGLM KVNNLHEYTK VPIFGCGLTT EMIIPSVNPS ICVEVPFDTE NNWAVTFDED AQRNYSWRDK G DTVTGHLV ...String: IKDILRRPVL LFNHVELDPD YTGFFIPIMP PSRMMQYKSG DKETSFQRLI GRTPQAAIMN LFRFWRGSLR YTIIIHSTDG HPIYVTHVP HTGNRVYGLM KVNNLHEYTK VPIFGCGLTT EMIIPSVNPS ICVEVPFDTE NNWAVTFDED AQRNYSWRDK G DTVTGHLV VTPVVSVYMS VWVEAGDDFE VSNFYGPPSV KTNDWNYAFS DEH UniProtKB: Genome polyprotein |
-Macromolecule #2: structural protein VP2
| Macromolecule | Name: structural protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sacbrood virus Sacbrood virus |
| Molecular weight | Theoretical: 20.360314 KDa |
| Sequence | String: WMPINSIRVT VNGKRNDLLA QYYIPEDFLS THAKCAPNTI PFETYVYGKY ELEMKFVANG NKFQCGKVII SVKFDSYQAD NINTGFQAA LSRPHIMLDL STNNEGVLKI PFRYHRAFVR NQTHKTATAG VRPGKFASIY VQVLSPLQTG EGGANDMFIR P FYRYTRAE FAGMSYKVPL T UniProtKB: Genome polyprotein |
-Macromolecule #3: structural protein VP3
| Macromolecule | Name: structural protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sacbrood virus Sacbrood virus |
| Molecular weight | Theoretical: 25.574461 KDa |
| Sequence | String: DEPRTTLDIA RIWGLRSTFN WGSGDEHGKE LFNTVLDPGL RFYDQDYEGQ ITPMEYVTGL YNFWSGPIEL RFDFVSNAFH TGTVIISAE YNRSSTNTDE CQSHSTYTKT FHLGEQKSVH FTVPYIYDTV VRRNTASAYL PVTDYDKVDN VSRAQAMGIR A ESKMRVKV ...String: DEPRTTLDIA RIWGLRSTFN WGSGDEHGKE LFNTVLDPGL RFYDQDYEGQ ITPMEYVTGL YNFWSGPIEL RFDFVSNAFH TGTVIISAE YNRSSTNTDE CQSHSTYTKT FHLGEQKSVH FTVPYIYDTV VRRNTASAYL PVTDYDKVDN VSRAQAMGIR A ESKMRVKV RVVNVLRPVA STTSTIEVLV YMRGGKNYAL HGLKQSTYWP SNSVVPIDSF PPDGYDP UniProtKB: Genome polyprotein |
-Macromolecule #4: minor capsid protein MiCP
| Macromolecule | Name: minor capsid protein MiCP / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sacbrood virus Sacbrood virus |
| Molecular weight | Theoretical: 3.107398 KDa |
| Sequence | String: DNPHRFLPAN VSNRWNEYSS AYLPRV UniProtKB: Structural protein Vp1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 101.325 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Preliminar grid screening was performed manually on FEI Tecnai F20 (cryo stage). |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 2-16 / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 21.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 74325 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 268.71 / Target criteria: R-factor |
|---|---|
| Output model |  PDB-6eh1: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)