[English] 日本語
 Yorodumi
Yorodumi- EMDB-3855: CryoEM structure of recombinant CMV particles with Tetanus-epitope -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3855 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of recombinant CMV particles with Tetanus-epitope | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | CryoEM structure of recombinant CMV particles with Tetanus-epitope != cucumber mosaic cucumovirus CryoEM structure of recombinant CMV particles with Tetanus-epitope
| |||||||||
 Keywords Keywords | VLP / Vaccines / CryoEM / CMV / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=3 icosahedral viral capsid / viral nucleocapsid / ribonucleoprotein complex / structural molecule activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Cucumber mosaic virus (cucumber mosaic cucumovirus) / Cucumber mosaic virus (cucumber mosaic cucumovirus) /  cucumber mosaic cucumovirus (cucumber mosaic cucumovirus) cucumber mosaic cucumovirus (cucumber mosaic cucumovirus) | |||||||||
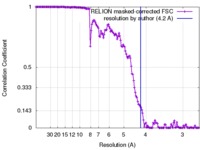
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Kotecha A / Stuart DI | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2017 Journal: NPJ Vaccines / Year: 2017Title: Incorporation of tetanus-epitope into virus-like particles achieves vaccine responses even in older recipients in models of psoriasis, Alzheimer's and cat allergy. Authors: Andris Zeltins / Jonathan West / Franziska Zabel / Aadil El Turabi / Ina Balke / Stefanie Haas / Melanie Maudrich / Federico Storni / Paul Engeroff / Gary T Jennings / Abhay Kotecha / David ...Authors: Andris Zeltins / Jonathan West / Franziska Zabel / Aadil El Turabi / Ina Balke / Stefanie Haas / Melanie Maudrich / Federico Storni / Paul Engeroff / Gary T Jennings / Abhay Kotecha / David I Stuart / John Foerster / Martin F Bachmann /    Abstract: Monoclonal antibodies are widely used to treat non-infectious conditions but are costly. Vaccines could offer a cost-effective alternative but have been limited by sub-optimal T-cell stimulation ...Monoclonal antibodies are widely used to treat non-infectious conditions but are costly. Vaccines could offer a cost-effective alternative but have been limited by sub-optimal T-cell stimulation and/or weak vaccine responses in recipients, for example, in elderly patients. We have previously shown that the repetitive structure of virus-like-particles (VLPs) can effectively bypass self-tolerance in therapeutic vaccines. Their efficacy could be increased even further by the incorporation of an epitope stimulating T cell help. However, the self-assembly and stability of VLPs from envelope monomer proteins is sensitive to geometry, rendering the incorporation of foreign epitopes difficult. We here show that it is possible to engineer VLPs derived from a non human-pathogenic plant virus to incorporate a powerful T-cell-stimulatory epitope derived from Tetanus toxoid. These VLPs (termed CMV) retain self-assembly as well as long-term stability. Since Th cell memory to Tetanus is near universal in humans, CMV-based vaccines can deliver robust antibody-responses even under limiting conditions. By way of proof of concept, we tested a range of such vaccines against chronic inflammatory conditions (model: psoriasis, antigen: interleukin-17), neurodegenerative (Alzheimer's, β-amyloid), and allergic disease (cat allergy, Fel-d1), respectively. Vaccine responses were uniformly strong, selective, efficient , observed even in old mice, and employing low vaccine doses. In addition, randomly ascertained human blood cells were reactive to CMV-VLPs, confirming recognition of the incorporated Tetanus epitope. The CMV-VLP platform is adaptable to almost any antigen and its features and performance are ideally suited for the design of vaccines delivering enhanced responsiveness in aging populations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3855.map.gz emd_3855.map.gz | 228.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3855-v30.xml emd-3855-v30.xml emd-3855.xml emd-3855.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3855_fsc.xml emd_3855_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_3855.png emd_3855.png | 238 KB | ||
| Filedesc metadata |  emd-3855.cif.gz emd-3855.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3855 http://ftp.pdbj.org/pub/emdb/structures/EMD-3855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3855 | HTTPS FTP |
-Validation report
| Summary document |  emd_3855_validation.pdf.gz emd_3855_validation.pdf.gz | 313.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3855_full_validation.pdf.gz emd_3855_full_validation.pdf.gz | 312.4 KB | Display | |
| Data in XML |  emd_3855_validation.xml.gz emd_3855_validation.xml.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3855 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3855 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3855 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3855 | HTTPS FTP |
-Related structure data
| Related structure data |  5ow6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3855.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3855.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CryoEM structure of recombinant CMV particles with Tetanus-epitope
| Entire | Name: CryoEM structure of recombinant CMV particles with Tetanus-epitope |
|---|---|
| Components |
|
-Supramolecule #1: cucumber mosaic cucumovirus
| Supramolecule | Name: cucumber mosaic cucumovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12305 / Sci species name: cucumber mosaic cucumovirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Cucumis sativus (cucumber) Cucumis sativus (cucumber) |
| Virus shell | Shell ID: 1 / T number (triangulation number): 1 |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 Details: PGYTFTSITLKPPKIDRGSYYGKRLLLPDSVTEYDKKLVSRLQIRVNPLPKFDSTVWVTVRKVPASSDLSVAAISAMFADGASPVLVYQYAASGVQANNKLLYDLSAMRADIGDMRKYAVLVYSKDDALETDELVLHVDIEHQRIPTSGVLPV Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cucumber mosaic virus (cucumber mosaic cucumovirus) Cucumber mosaic virus (cucumber mosaic cucumovirus) |
| Molecular weight | Theoretical: 16.92442 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PGYTFTSITL KPPKIDRGSY YGKRLLLPDS VTEYDKKLVS RLQIRVNPLP KFDSTVWVTV RKVPASSDLS VAAISAMFAD GASPVLVYQ YAASGVQANN KLLYDLSAMR ADIGDMRKYA VLVYSKDDAL ETDELVLHVD IEHQRIPTSG VLPV UniProtKB: Capsid protein |
-Macromolecule #2: Capsid protein, VP2
| Macromolecule | Name: Capsid protein, VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cucumber mosaic virus (cucumber mosaic cucumovirus) Cucumber mosaic virus (cucumber mosaic cucumovirus) |
| Molecular weight | Theoretical: 21.068152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DANFRVLSQQ LSRLNKTLAA GRPTINHPTF VGSERCRPGY TFTSITLKPP KIDRGSYYGK RLLLPDSVTE YDKKLVSRLQ IRVNPLPKF DSTVWVTVRK VPASSDLSVA AISAMFADGA SPVLVYQYAA SGVQANNKLL YDLSAMRADI GDMRKYAVLV Y SKDDALET DELVLHVDIE HQRIPTSGVL PV UniProtKB: Capsid protein |
-Macromolecule #3: Capsid protein, VP3
| Macromolecule | Name: Capsid protein, VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Cucumber mosaic virus (cucumber mosaic cucumovirus) Cucumber mosaic virus (cucumber mosaic cucumovirus) |
| Molecular weight | Theoretical: 21.139229 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ADANFRVLSQ QLSRLNKTLA AGRPTINHPT FVGSERCRPG YTFTSITLKP PKIDRGSYYG KRLLLPDSVT EYDKKLVSRL QIRVNPLPK FDSTVWVTVR KVPASSDLSV AAISAMFADG ASPVLVYQYA ASGVQANNKL LYDLSAMRAD IGDMRKYAVL V YSKDDALE TDELVLHVDI EHQRIPTSGV LPV UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 9 / Details: 5 mM sodium borate, 2 mM EDTA, pH 9 |
| Grid | Model: C Flats 2/1 2C / Material: COPPER / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 17 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | 5 mM sodium borate, 2 mM EDTA, pH 9 |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Energy filter - Name: GIF / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-25 / Number grids imaged: 1 / Number real images: 500 / Average exposure time: 5.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 160000 |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 174 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5ow6: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)