[English] 日本語
 Yorodumi
Yorodumi- EMDB-38432: Core region of the citrate-induced human acetyl-CoA carboxylase 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Biotin-dependent carboxylase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetyl-CoA carboxylase / fatty-acyl-CoA biosynthetic process / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / malonyl-CoA biosynthetic process / Fatty acyl-CoA biosynthesis / ChREBP activates metabolic gene expression / acetyl-CoA carboxylase activity / acetyl-CoA metabolic process / tissue homeostasis ...acetyl-CoA carboxylase / fatty-acyl-CoA biosynthetic process / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / malonyl-CoA biosynthetic process / Fatty acyl-CoA biosynthesis / ChREBP activates metabolic gene expression / acetyl-CoA carboxylase activity / acetyl-CoA metabolic process / tissue homeostasis / Carnitine shuttle / lipid homeostasis / Activation of gene expression by SREBF (SREBP) / fibrillar center / fatty acid biosynthetic process / cellular response to prostaglandin E stimulus / actin cytoskeleton / protein homotetramerization / mitochondrion / ATP binding / metal ion binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Zhou FY / Zhang YY / Zhou Q / Hu Q | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase. Authors: Fayang Zhou / Yuanyuan Zhang / Yuyao Zhu / Qiang Zhou / Yigong Shi / Qi Hu /  Abstract: Human acetyl-coenzyme A (CoA) carboxylases (ACCs) catalyze the carboxylation of acetyl-CoA, which is the rate-limiting step in fatty acid synthesis. The molecular mechanism underlying the dynamic ...Human acetyl-coenzyme A (CoA) carboxylases (ACCs) catalyze the carboxylation of acetyl-CoA, which is the rate-limiting step in fatty acid synthesis. The molecular mechanism underlying the dynamic organization of ACCs is largely unknown. Here, we determined the cryo-electron microscopy (EM) structure of human ACC1 in its inactive state, which forms a unique filament structure and is in complex with acetyl-CoA. We also determined the cryo-EM structure of human ACC1 activated by dephosphorylation and citrate treatment, at a resolution of 2.55 Å. Notably, the covalently linked biotin binds to a site that is distant from the acetyl-CoA binding site when acetyl-CoA is absent, suggesting a potential coordination between biotin binding and acetyl-CoA binding. These findings provide insights into the structural dynamics and regulatory mechanisms of human ACCs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38432.map.gz emd_38432.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38432-v30.xml emd-38432-v30.xml emd-38432.xml emd-38432.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38432.png emd_38432.png | 38 KB | ||
| Filedesc metadata |  emd-38432.cif.gz emd-38432.cif.gz | 6.6 KB | ||
| Others |  emd_38432_half_map_1.map.gz emd_38432_half_map_1.map.gz emd_38432_half_map_2.map.gz emd_38432_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38432 http://ftp.pdbj.org/pub/emdb/structures/EMD-38432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38432 | HTTPS FTP |
-Validation report
| Summary document |  emd_38432_validation.pdf.gz emd_38432_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38432_full_validation.pdf.gz emd_38432_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_38432_validation.xml.gz emd_38432_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_38432_validation.cif.gz emd_38432_validation.cif.gz | 16.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38432 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38432 | HTTPS FTP |
-Related structure data
| Related structure data |  8xkzMC  8xl0C  8xl1C  8xl2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38432.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38432.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||||||||||||||||||
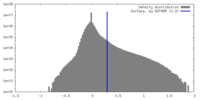
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38432_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
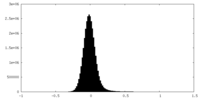
| Density Histograms |
-Half map: #1
| File | emd_38432_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Core region of the citrate-induced human acetyl-CoA carboxylase 1...
| Entire | Name: Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) |
|---|---|
| Components |
|
-Supramolecule #1: Core region of the citrate-induced human acetyl-CoA carboxylase 1...
| Supramolecule | Name: Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Acetyl-CoA carboxylase 1
| Macromolecule | Name: Acetyl-CoA carboxylase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: acetyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 254.443312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RDFTVASPAE FVTRFGGNKV IEKVLIANNG IAAVKCMRSI RRWSYEMFRN ERAIRFVVMV TPEDLKANAE YIKMADHYVP VPGGPNNNN YANVELILDI AKRIPVQAVW AGWGHASENP KLPELLLKNG IAFMGPPSQA MWALGDKIAS SIVAQTAGIP T LPWSGSGL ...String: RDFTVASPAE FVTRFGGNKV IEKVLIANNG IAAVKCMRSI RRWSYEMFRN ERAIRFVVMV TPEDLKANAE YIKMADHYVP VPGGPNNNN YANVELILDI AKRIPVQAVW AGWGHASENP KLPELLLKNG IAFMGPPSQA MWALGDKIAS SIVAQTAGIP T LPWSGSGL RVDWQENDFS KRILNVPQEL YEKGYVKDVD DGLQAAEEVG YPVMIKASEG GGGKGIRKVN NADDFPNLFR QV QAEVPGS PIFVMRLAKQ SRHLEVQILA DQYGNAISLF GRDCSVQRRH QKIIEEAPAT IATPAVFEHM EQCAVKLAKM VGY VSAGTV EYLYSQDGSF YFLELNPRLQ VEHPCTEMVA DVNLPAAQLQ IAMGIPLYRI KDIRMMYGVS PWGDSPIDFE DSAH VPCPR GHVIAARITS ENPDEGFKPS SGTVQELNFR SNKNVWGYFS VAAAGGLHEF ADSQFGHCFS WGENREEAIS NMVVA LKEL SIRGDFRTTV EYLIKLLETE SFQMNRIDTG WLDRLIAEKV QAERPDTMLG VVCGALHVAD VSLRNSVSNF LHSLER GQV LPAHTLLNTV DVELIYEGVK YVLKVTRQSP NSYVVIMNGS CVEVDVHRLS DGGLLLSYDG SSYTTYMKEE VDRYRIT IG NKTCVFEKEN DPSVMRSPSA GKLIQYIVED GGHVFAGQCY AEIEVMKMVM TLTAVESGCI HYVKRPGAAL DPGCVLAK M QLDNPSKVQQ AELHTGSLPR IQSTALRGEK LHRVFHYVLD NLVNVMNGYC LPDPFFSSKV KDWVERLMKT LRDPSLPLL ELQDIMTSVS GRIPPNVEKS IKKEMAQYAS NITSVLCQFP SQQIANILDS HAATLNRKSE REVFFMNTQS IVQLVQRYRS GIRGHMKAV VMDLLRQYLR VETQFQNGHY DKCVFALREE NKSDMNTVLN YIFSHAQVTK KNLLVTMLID QLCGRDPTLT D ELLNILTE LTQLSKTTNA KVALRARQVL IASHLPSYEL RHNQVESIFL SAIDMYGHQF CIENLQKLIL SETSIFDVLP NF FYHSNQV VRMAALEVYV RRAYIAYELN SVQHRQLKDN TCVVEFQFML PTSHPNRGNI PTLNRMSFSS NLNHYGMTHV ASV SDVLLD NSFTPPCQRM GGMVSFRTFE DFVRIFDEVM GCFSDSPPQS PTFPEAGHTS LYDEDKVPRD EPIHILNVAI KTDC DIEDD RLAAMFREFT QQNKATLVDH GIRRLTFLVA QKDFRKQVNY EVDRRFHREF PKFFTFRARD KFEEDRIYRH LEPAL AFQL ELNRMRNFDL TAIPCANHKM HLYLGAAKVE VGTEVTDYRF FVRAIIRHSD LVTKEASFEY LQNEGERLLL EAMDEL EVA FNNTNVRTDC NHIFLNFVPT VIMDPSKIEE SVRSMVMRYG SRLWKLRVLQ AELKINIRLT PTGKAIPIRL FLTNESG YY LDISLYKEVT DSRTAQIMFQ AYGDKQGPLH GMLINTPYVT KDLLQSKRFQ AQSLGTTYIY DIPEMFRQSL IKLWESMS T QAFLPSPPLP SDMLTYTELV LDDQGQLVHM NRLPGGNEIG MVAWKMTFKS PEYPEGRDII VIGNDITYRI GSFGPQEDL LFLRASELAR AEGIPRIYVS ANSGARIGLA EEIRHMFHVA WVDPEDPYKG YRYLYLTPQD YKRVSALNSV HCEHVEDEGE SRYKITDII GKEEGIGPEN LRGSGMIAGE SSLAYNEIIT ISLVTCRAIG IGAYLVRLGQ RTIQVENSHL ILTGAGALNK V LGREVYTS NNQLGGIQIM HNNGVTHCTV CDDFEGVFTV LHWLSYMPKS VHSSVPLLNS KDPIDRIIEF VPTKTPYDPR WM LAGRPHP TQKGQWLSGF FDYGSFSEIM QPWAQTVVVG RARLGGIPVG VVAVETRTVE LSIPADPANL DSEAKIIQQA GQV WFPDSA FKTYQAIKDF NREGLPLMVF ANWRGFSGGM KDMYDQVLKF GAYIVDGLRE CCQPVLVYIP PQAELRGGSW VVID SSINP RHMEMYADRE SRGSVLEPEG TVEIKFRRKD LVKTMRRVDP VYIHLAERLG TPELSTAERK ELENKLKERE EFLIP IYHQ VAVQFADLHD TPGRMQEKGV ISDILDWKTS RTFFYWRLRR LLLEDLVKKK IHNANPELTD GQIQAMLRRW FVEVEG TVK AYVWDNNKDL AEWLEKQLTE EDGVHSVIEE NIKCISRDYV LKQIRSLVQA NPEVAMDSII HMTQHISPTQ RAEVIRI UniProtKB: Acetyl-CoA carboxylase 1 |
-Macromolecule #2: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 2 / Number of copies: 2 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information |  ChemComp-BTN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.55 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 224904 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)