[English] 日本語
 Yorodumi
Yorodumi- EMDB-38344: Human Cx36/GJD2 (Ala14-deleted mutant) gap junction channel in po... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Cx36/GJD2 (Ala14-deleted mutant) gap junction channel in porcine brain lipids | |||||||||
 Map data Map data | Cryo-EM density map, D6 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | connexin 36 / Cx36 / Gap Junction Channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions / connexin complex / Gap junction assembly / gap junction channel activity / neuronal action potential / visual perception / cell-cell signaling / chemical synaptic transmission / synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.72 Å | |||||||||
 Authors Authors | Cho HJ / Lee HH | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer. Authors: Hwa-Jin Cho / Dong Kyu Chung / Hyung Ho Lee /  Abstract: Connexin 36 (Cx36) forms interneuronal gap junctions, establishing electrical synapses for rapid synaptic transmission. In disease conditions, inhibiting Cx36 gap junction channels (GJCs) is ...Connexin 36 (Cx36) forms interneuronal gap junctions, establishing electrical synapses for rapid synaptic transmission. In disease conditions, inhibiting Cx36 gap junction channels (GJCs) is beneficial, as it prevents abnormal synchronous neuronal firing and apoptotic signal propagation, mitigating seizures and progressive cell death. Here, we present cryo-electron microscopy structures of human Cx36 GJC in complex with known channel inhibitors, such as mefloquine, arachidonic acid, and 1-hexanol. Notably, these inhibitors competitively bind to the binding pocket of the N-terminal helices (NTH), inducing a conformational shift from the pore-lining NTH (PLN) state to the flexible NTH (FN) state. This leads to the obstruction of the channel pore by flat double-layer densities of lipids. These studies elucidate the molecular mechanisms of how Cx36 GJC can be modulated by inhibitors, providing valuable insights into potential therapeutic applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38344.map.gz emd_38344.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38344-v30.xml emd-38344-v30.xml emd-38344.xml emd-38344.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38344_fsc.xml emd_38344_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_38344.png emd_38344.png | 123 KB | ||
| Filedesc metadata |  emd-38344.cif.gz emd-38344.cif.gz | 5.9 KB | ||
| Others |  emd_38344_half_map_1.map.gz emd_38344_half_map_1.map.gz emd_38344_half_map_2.map.gz emd_38344_half_map_2.map.gz | 115.8 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38344 http://ftp.pdbj.org/pub/emdb/structures/EMD-38344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38344 | HTTPS FTP |
-Related structure data
| Related structure data |  8xh8MC  8xgdC  8xgeC  8xgfC  8xggC  8xgjC  8xh9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38344.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38344.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM density map, D6 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.858 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_38344_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_38344_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Cx36/GJD2 gap junction channel
| Entire | Name: Human Cx36/GJD2 gap junction channel |
|---|---|
| Components |
|
-Supramolecule #1: Human Cx36/GJD2 gap junction channel
| Supramolecule | Name: Human Cx36/GJD2 gap junction channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction delta-2 protein
| Macromolecule | Name: Gap junction delta-2 protein / type: protein_or_peptide / ID: 1 Details: Residues M1-V320: connexin 36 (Ala14-deleted mutant) Residues S321-R322: linker Residues D323-K330: affinity tag (FLAG) Following regions are not modeled because of ambiguity of density map: ...Details: Residues M1-V320: connexin 36 (Ala14-deleted mutant) Residues S321-R322: linker Residues D323-K330: affinity tag (FLAG) Following regions are not modeled because of ambiguity of density map: Residues M1-S19, S100-I194, and W276-K330) Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.305832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGEWTILERL LEAVQQHSTM IGRILLTVVV IFRILIVAIV GETVYDDEQT MFVCNTLQPG CNQACYDRAF PISHIRYWVF QIIMVCTPS LCFITYSVHQ SAKQRERRYS TVFLALDRDP PESIGGPGGT GGGGSGGGKR EDKKLQNAIV NGVLQNTENT S KETEPDCL ...String: MGEWTILERL LEAVQQHSTM IGRILLTVVV IFRILIVAIV GETVYDDEQT MFVCNTLQPG CNQACYDRAF PISHIRYWVF QIIMVCTPS LCFITYSVHQ SAKQRERRYS TVFLALDRDP PESIGGPGGT GGGGSGGGKR EDKKLQNAIV NGVLQNTENT S KETEPDCL EVKELTPHPS GLRTASKSKL RRQEGISRFY IIQVVFRNAL EIGFLVGQYF LYGFSVPGLY ECNRYPCIKE VE CYVSRPT EKTVFLVFMF AVSGICVVLN LAELNHLGWR KIKLAVRGAQ AKRKSIYEIR NKDLPRVSVP NFGRTQSSDS AYV SRDYKD DDDK UniProtKB: Gap junction delta-2 protein |
-Macromolecule #2: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 72 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information | 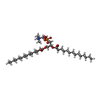 ChemComp-MC3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































