+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3834 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an unphosphorylated MCM double hexamer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
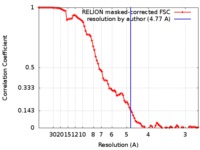
| Method | single particle reconstruction / cryo EM / Resolution: 4.77 Å | |||||||||
 Authors Authors | Abid Ali F | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Cryo-EM structure of a licensed DNA replication origin. Authors: Ferdos Abid Ali / Max E Douglas / Julia Locke / Valerie E Pye / Andrea Nans / John F X Diffley / Alessandro Costa /  Abstract: Eukaryotic origins of replication are licensed upon loading of the MCM helicase motor onto DNA. ATP hydrolysis by MCM is required for loading and the post-catalytic MCM is an inactive double hexamer ...Eukaryotic origins of replication are licensed upon loading of the MCM helicase motor onto DNA. ATP hydrolysis by MCM is required for loading and the post-catalytic MCM is an inactive double hexamer that encircles duplex DNA. Origin firing depends on MCM engagement of Cdc45 and GINS to form the CMG holo-helicase. CMG assembly requires several steps including MCM phosphorylation by DDK. To understand origin activation, here we have determined the cryo-EM structures of DNA-bound MCM, either unmodified or phosphorylated, and visualize a phospho-dependent MCM element likely important for Cdc45 recruitment. MCM pore loops touch both the Watson and Crick strands, constraining duplex DNA in a bent configuration. By comparing our new MCM-DNA structure with the structure of CMG-DNA, we suggest how the conformational transition from the loaded, post-catalytic MCM to CMG might promote DNA untwisting and melting at the onset of replication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3834.map.gz emd_3834.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3834-v30.xml emd-3834-v30.xml emd-3834.xml emd-3834.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3834_fsc.xml emd_3834_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_3834.png emd_3834.png | 71.8 KB | ||
| Others |  emd_3834_half_map_1.map.gz emd_3834_half_map_1.map.gz emd_3834_half_map_2.map.gz emd_3834_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3834 http://ftp.pdbj.org/pub/emdb/structures/EMD-3834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3834 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3834.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3834.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
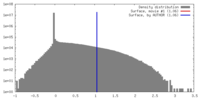
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_3834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_3834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae unphosphorylated MCM double hexamer
| Entire | Name: Saccharomyces cerevisiae unphosphorylated MCM double hexamer |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae unphosphorylated MCM double hexamer
| Supramolecule | Name: Saccharomyces cerevisiae unphosphorylated MCM double hexamer type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.2 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.67 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)