+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | consensus map of RS head-neck monomer complex | |||||||||
 Map data Map data | sharpened consensus map of RS head-neck monomer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Radial spoke / cilia / axoneme / STRUCTURAL PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Meng X / Cong Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia. Authors: Xueming Meng / Cong Xu / Jiawei Li / Benhua Qiu / Jiajun Luo / Qin Hong / Yujie Tong / Chuyu Fang / Yanyan Feng / Rui Ma / Xiangyi Shi / Cheng Lin / Chen Pan / Xueliang Zhu / Xiumin Yan / Yao Cong /  Abstract: Radial spokes (RS) transmit mechanochemical signals between the central pair (CP) and axonemal dynein arms to coordinate ciliary motility. Atomic-resolution structures of metazoan RS and structures ...Radial spokes (RS) transmit mechanochemical signals between the central pair (CP) and axonemal dynein arms to coordinate ciliary motility. Atomic-resolution structures of metazoan RS and structures of axonemal complexes in ependymal cilia, whose rhythmic beating drives the circulation of cerebrospinal fluid, however, remain obscure. Here, we present near-atomic resolution cryo-EM structures of mouse RS head-neck complex in both monomer and dimer forms and reveal the intrinsic flexibility of the dimer. We also map the genetic mutations related to primary ciliary dyskinesia and asthenospermia on the head-neck complex. Moreover, we present the cryo-ET and sub-tomogram averaging map of mouse ependymal cilia and build the models for RS1-3, IDAs, and N-DRC. Contrary to the conserved RS structure, our cryo-ET map reveals the lack of IDA-b/c/e and the absence of Tektin filaments within the A-tubule of doublet microtubules in ependymal cilia compared with mammalian respiratory cilia and sperm flagella, further exemplifying the structural diversity of mammalian motile cilia. Our findings shed light on the stepwise mammalian RS assembly mechanism, the coordinated rigid and elastic RS-CP interaction modes beneficial for the regulation of asymmetric ciliary beating, and also facilitate understanding on the etiology of ciliary dyskinesia-related ciliopathies and on the ependymal cilia in the development of hydrocephalus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38004.map.gz emd_38004.map.gz | 584.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38004-v30.xml emd-38004-v30.xml emd-38004.xml emd-38004.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38004.png emd_38004.png | 56.8 KB | ||
| Filedesc metadata |  emd-38004.cif.gz emd-38004.cif.gz | 3.9 KB | ||
| Others |  emd_38004_additional_1.map.gz emd_38004_additional_1.map.gz emd_38004_half_map_1.map.gz emd_38004_half_map_1.map.gz emd_38004_half_map_2.map.gz emd_38004_half_map_2.map.gz | 335.1 MB 620.9 MB 620.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38004 http://ftp.pdbj.org/pub/emdb/structures/EMD-38004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38004 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38004.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38004.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened consensus map of RS head-neck monomer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.854 Å | ||||||||||||||||||||||||||||||||||||
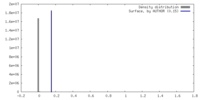
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpening consensus map of RS head-neck monomer
| File | emd_38004_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpening consensus map of RS head-neck monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
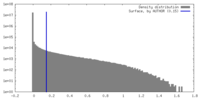
| Density Histograms |
-Half map: half map A for consensus map of RS head-neck monomer
| File | emd_38004_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A for consensus map of RS head-neck monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
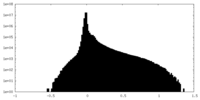
| Density Histograms |
-Half map: half map B for consensus map of RS head-neck monomer
| File | emd_38004_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B for consensus map of RS head-neck monomer | ||||||||||||
| Projections & Slices |
| ||||||||||||
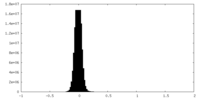
| Density Histograms |
- Sample components
Sample components
-Entire : RS head-neck complex
| Entire | Name: RS head-neck complex |
|---|---|
| Components |
|
-Supramolecule #1: RS head-neck complex
| Supramolecule | Name: RS head-neck complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.28 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 433940 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)