[English] 日本語
 Yorodumi
Yorodumi- EMDB-37900: Cryo-EM structure of human SLC15A4 in complex with Lys-Leu (outwa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human SLC15A4 in complex with Lys-Leu (outward-facing open) | |||||||||
 Map data Map data | B-factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhistidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / L-histidine transmembrane transporter activity / peptidoglycan transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes ...histidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / L-histidine transmembrane transporter activity / peptidoglycan transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes / SLC15A4:TASL-dependent IRF5 activation / positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway / dipeptide import across plasma membrane / positive regulation of toll-like receptor 9 signaling pathway / peptide:proton symporter activity / dipeptide transmembrane transporter activity / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / endolysosome membrane / positive regulation of innate immune response / specific granule membrane / monoatomic ion transport / protein transport / early endosome membrane / innate immune response / lysosomal membrane / Neutrophil degranulation / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Sakaniwa K / Zhang Z / Ohto U / Shimizu T | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: The structures of the peptide transporters SLC15A3 and SLC15A4 reveal the recognition mechanisms for substrate and TASL. Authors: Zhikuan Zhang / Shota Kasai / Kentaro Sakaniwa / Akiko Fujimura / Umeharu Ohto / Toshiyuki Shimizu /  Abstract: The solute carrier family 15 members 3 and 4 (SLC15A3 and SLC15A4) are closely related endolysosomal peptide transporters that transport free histidine and certain dipeptides from the lumen to ...The solute carrier family 15 members 3 and 4 (SLC15A3 and SLC15A4) are closely related endolysosomal peptide transporters that transport free histidine and certain dipeptides from the lumen to cytosol. Besides, SLC15A4 also functions as a scaffold protein for the recruitment of the adapter TASL for interferon regulatory factor 5 (IRF5) activation downstream of innate immune TLR7-9 signaling. However, the molecular basis for the substrate recognition and TASL recruitment by these membrane proteins is not well understood. Here, we report the cryoelectron microscopy (cryo-EM) structure of apo SLC15A3 and structures of SLC15A4 in the absence or presence of the substrate, revealing the specific dipeptide recognition mechanism. Each SLC15A3 and SLC15A4 protomer adopts an outward-facing conformation. Furthermore, we also present the cryo-EM structure of a SLC15A4-TASL complex. The N terminal region of TASL forms a helical structure that inserts deeply into the inward-facing cavity of SLC15A4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37900.map.gz emd_37900.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37900-v30.xml emd-37900-v30.xml emd-37900.xml emd-37900.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37900.png emd_37900.png | 116.2 KB | ||
| Filedesc metadata |  emd-37900.cif.gz emd-37900.cif.gz | 6.1 KB | ||
| Others |  emd_37900_additional_1.map.gz emd_37900_additional_1.map.gz emd_37900_half_map_1.map.gz emd_37900_half_map_1.map.gz emd_37900_half_map_2.map.gz emd_37900_half_map_2.map.gz | 15.2 MB 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37900 http://ftp.pdbj.org/pub/emdb/structures/EMD-37900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37900 | HTTPS FTP |
-Related structure data
| Related structure data |  8wx4MC  8wx1C  8wx2C  8wx3C  8wx5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37900.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37900.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
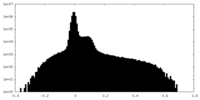
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_37900_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
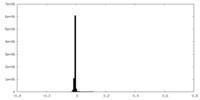
| Density Histograms |
-Half map: #1
| File | emd_37900_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
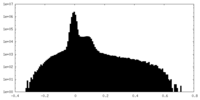
| Density Histograms |
-Half map: #2
| File | emd_37900_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
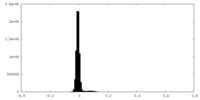
| Density Histograms |
- Sample components
Sample components
-Entire : Human SLC15A4 with KL
| Entire | Name: Human SLC15A4 with KL |
|---|---|
| Components |
|
-Supramolecule #1: Human SLC15A4 with KL
| Supramolecule | Name: Human SLC15A4 with KL / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 15 member 4
| Macromolecule | Name: Solute carrier family 15 member 4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 64.856176 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGSGGGAGE RAPLLGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ...String: MEGSGGGAGE RAPLLGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ATVKANITPF GADQVKDRGP EATRRFFNWF YWSINLGAIL SLGGIAYIQQ NVSFVTGYAI PTVCVGLAFV VF LCGQSVF ITKPPDGSAF TDMFKILTYS CCSQKRSGER QSNGEGIGVF QQSSKQSLFD SCKMSHGGPF TEEKVEDVKA LVK IVPVFL ALIPYWTVYF QMQTTYVLQS LHLRIPEISN ITTTPHTLPA AWLTMFDAVL ILLLIPLKDK LVDPILRRHG LLPS SLKRI AVGMFFVMCS AFAAGILESK RLNLVKEKTI NQTIGNVVYH AADLSLWWQV PQYLLIGISE IFASIAGLEF AYSAA PKSM QSAIMGLFFF FSGVGSFVGS GLLALVSIKA IGWMSSHTDF GNINGCYLNY YFFLLAAIQG ATLLLFLIIS VKYDHH RDH QRSRANGVPT SRRAGSIEGR IVKDYKDDDD KHHHHHH UniProtKB: Solute carrier family 15 member 4 |
-Macromolecule #2: LYSINE
| Macromolecule | Name: LYSINE / type: ligand / ID: 2 / Number of copies: 2 / Formula: LYS |
|---|---|
| Molecular weight | Theoretical: 147.195 Da |
| Chemical component information |  ChemComp-LYS: |
-Macromolecule #3: LEUCINE
| Macromolecule | Name: LEUCINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: LEU |
|---|---|
| Molecular weight | Theoretical: 131.173 Da |
| Chemical component information |  ChemComp-LEU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)