[English] 日本語
 Yorodumi
Yorodumi- EMDB-37565: Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH. | |||||||||
 Map data Map data | Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / protein synthesis / Mycobacterium smegmatis / hibernation promotion factor / HPF / RafH / hypoxia stress / Cryo- EM / Single particle reconstruction | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of translational elongation / ribosomal small subunit binding / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome ...negative regulation of translational elongation / ribosomal small subunit binding / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
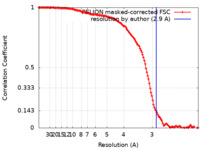
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Kumar N / Sharma S / Kaushal PS | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH. Authors: Niraj Kumar / Shivani Sharma / Prem S Kaushal /  Abstract: Ribosome hibernation is a key survival strategy bacteria adopt under environmental stress, where a protein, hibernation promotion factor (HPF), transitorily inactivates the ribosome. Mycobacterium ...Ribosome hibernation is a key survival strategy bacteria adopt under environmental stress, where a protein, hibernation promotion factor (HPF), transitorily inactivates the ribosome. Mycobacterium tuberculosis encounters hypoxia (low oxygen) as a major stress in the host macrophages, and upregulates the expression of RafH protein, which is crucial for its survival. The RafH, a dual domain HPF, an orthologue of bacterial long HPF (HPF), hibernates ribosome in 70S monosome form, whereas in other bacteria, the HPF induces 70S ribosome dimerization and hibernates its ribosome in 100S disome form. Here, we report the cryo- EM structure of M. smegmatis, a close homolog of M. tuberculosis, 70S ribosome in complex with the RafH factor at an overall 2.8 Å resolution. The N- terminus domain (NTD) of RafH binds to the decoding center, similarly to HPF NTD. In contrast, the C- terminus domain (CTD) of RafH, which is larger than the HPF CTD, binds to a distinct site at the platform binding center of the ribosomal small subunit. The two domain-connecting linker regions, which remain mostly disordered in earlier reported HPF structures, interact mainly with the anti-Shine Dalgarno sequence of the 16S rRNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37565.map.gz emd_37565.map.gz | 17.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37565-v30.xml emd-37565-v30.xml emd-37565.xml emd-37565.xml | 44.5 KB 44.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37565_fsc.xml emd_37565_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37565.png emd_37565.png | 95.1 KB | ||
| Filedesc metadata |  emd-37565.cif.gz emd-37565.cif.gz | 9.8 KB | ||
| Others |  emd_37565_half_map_1.map.gz emd_37565_half_map_1.map.gz emd_37565_half_map_2.map.gz emd_37565_half_map_2.map.gz | 140 MB 139.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37565 http://ftp.pdbj.org/pub/emdb/structures/EMD-37565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37565 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37565 | HTTPS FTP |
-Validation report
| Summary document |  emd_37565_validation.pdf.gz emd_37565_validation.pdf.gz | 703.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37565_full_validation.pdf.gz emd_37565_full_validation.pdf.gz | 703.2 KB | Display | |
| Data in XML |  emd_37565_validation.xml.gz emd_37565_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_37565_validation.cif.gz emd_37565_validation.cif.gz | 27.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37565 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37565 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37565 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37565 | HTTPS FTP |
-Related structure data
| Related structure data |  8wifMC  8whxC  8whyC  8wi7C  8wi8C  8wi9C  8wibC  8wicC  8widC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37565.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37565.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Mycobacterium smegmatis 30S ribosomal subunit (body 2 half...
| File | emd_37565_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mycobacterium smegmatis 30S ribosomal subunit (body 2 half 1 map) of 70S ribosome and RafH. | ||||||||||||
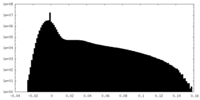
| Projections & Slices |
| ||||||||||||
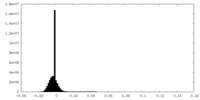
| Density Histograms |
-Half map: Mycobacterium smegmatis 30S ribosomal subunit (body 2 half...
| File | emd_37565_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mycobacterium smegmatis 30S ribosomal subunit (body 2 half 2 map) of 70S ribosome and RafH. | ||||||||||||
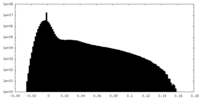
| Projections & Slices |
| ||||||||||||
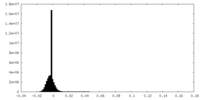
| Density Histograms |
- Sample components
Sample components
+Entire : 70S ribosome + RafH protein
+Supramolecule #1: 70S ribosome + RafH protein
+Supramolecule #2: 70S ribsome
+Supramolecule #3: RafH
+Macromolecule #1: 16S rRNA
+Macromolecule #2: 30S ribosomal protein S22
+Macromolecule #3: 30S ribosomal protein S3
+Macromolecule #4: 30S ribosomal protein S4
+Macromolecule #5: 30S ribosomal protein S5
+Macromolecule #6: 30S ribosomal protein S6
+Macromolecule #7: 30S ribosomal protein S7
+Macromolecule #8: 30S ribosomal protein S8
+Macromolecule #9: 30S ribosomal protein S9
+Macromolecule #10: 30S ribosomal protein S10
+Macromolecule #11: 30S ribosomal protein S11
+Macromolecule #12: 30S ribosomal protein S12
+Macromolecule #13: 30S ribosomal protein S13
+Macromolecule #14: 30S ribosomal protein S14A
+Macromolecule #15: 30S ribosomal protein S15
+Macromolecule #16: 30S ribosomal protein S16
+Macromolecule #17: 30S ribosomal protein S17
+Macromolecule #18: 30S ribosomal protein S18B
+Macromolecule #19: 30S ribosomal protein S19
+Macromolecule #20: 30S ribosomal protein S20
+Macromolecule #21: 30S ribosomal protein S2
+Macromolecule #22: 50S ribosomal protein L31
+Macromolecule #23: Ribosome hibernation promotion factor RafH
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20mM HEPES, 100mM NH4Cl, 20mM MgCl2, 3mM DTT, |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | 70S ribosome + RafH protein |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 3 / Number real images: 12343 / Average exposure time: 2.0 sec. / Average electron dose: 1.34 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | phenix.real_space_refinement |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8wif: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)