+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA BINDING PROTEIN-DNA COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Thermus thermophilus (bacteria) / Thermus thermophilus (bacteria) /   Thermococcus thioreducens (archaea) Thermococcus thioreducens (archaea) | |||||||||
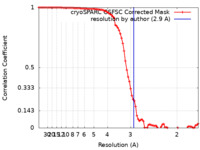
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Zhuang L | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute. Authors: Longyu Wang / Wanping Chen / Chendi Zhang / Xiaochen Xie / Fuyong Huang / Miaomiao Chen / Wuxiang Mao / Na Yu / Qiang Wei / Lixin Ma / Zhuang Li /  Abstract: The Argonaute nuclease from the thermophilic archaeon Pyrococcus furiosus (PfAgo) contributes to host defense and represents a promising biotechnology tool. Here, we report the structure of a PfAgo- ...The Argonaute nuclease from the thermophilic archaeon Pyrococcus furiosus (PfAgo) contributes to host defense and represents a promising biotechnology tool. Here, we report the structure of a PfAgo-guide DNA-target DNA ternary complex at the cleavage-compatible state. The ternary complex is predominantly dimerized, and the dimerization is solely mediated by PfAgo at PIWI-MID, PIWI-PIWI, and PAZ-N interfaces. Additionally, PfAgo accommodates a short 14-bp guide-target DNA duplex with a wedge-type N domain and specifically recognizes 5'-phosphorylated guide DNA. In contrast, the PfAgo-guide DNA binary complex is monomeric, and the engagement of target DNA with 14-bp complementarity induces sufficient dimerization and activation of PfAgo, accompanied by movement of PAZ and N domains. A closely related Argonaute from Thermococcus thioreducens adopts a similar dimerization configuration with an additional zinc finger formed at the dimerization interface. Dimerization of both Argonautes stabilizes the catalytic loops, highlighting the important role of Argonaute dimerization in the activation and target cleavage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37457.map.gz emd_37457.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37457-v30.xml emd-37457-v30.xml emd-37457.xml emd-37457.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37457_fsc.xml emd_37457_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37457.png emd_37457.png | 88.3 KB | ||
| Filedesc metadata |  emd-37457.cif.gz emd-37457.cif.gz | 6.8 KB | ||
| Others |  emd_37457_half_map_1.map.gz emd_37457_half_map_1.map.gz emd_37457_half_map_2.map.gz emd_37457_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37457 http://ftp.pdbj.org/pub/emdb/structures/EMD-37457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37457 | HTTPS FTP |
-Validation report
| Summary document |  emd_37457_validation.pdf.gz emd_37457_validation.pdf.gz | 963.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37457_full_validation.pdf.gz emd_37457_full_validation.pdf.gz | 963.3 KB | Display | |
| Data in XML |  emd_37457_validation.xml.gz emd_37457_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_37457_validation.cif.gz emd_37457_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37457 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37457 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37457 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37457 | HTTPS FTP |
-Related structure data
| Related structure data |  8wd8MC  8jpxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37457.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37457.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
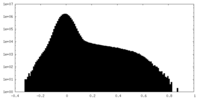
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37457_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
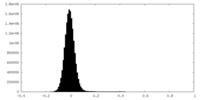
| Density Histograms |
-Half map: #1
| File | emd_37457_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
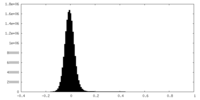
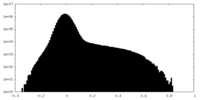
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of TtdAgo-guide DNA-target DNA complex
| Entire | Name: Cryo-EM structure of TtdAgo-guide DNA-target DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of TtdAgo-guide DNA-target DNA complex
| Supramolecule | Name: Cryo-EM structure of TtdAgo-guide DNA-target DNA complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Supramolecule #2: DNA binding protein
| Supramolecule | Name: DNA binding protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|
-Macromolecule #1: Argonaute family protein
| Macromolecule | Name: Argonaute family protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermococcus thioreducens (archaea) Thermococcus thioreducens (archaea) |
| Molecular weight | Theoretical: 87.488398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLMKVLTNMV KLNQDIIPNE IYLYKIFNKP EDGMNIYKIA YRNHGIVIDP QNRIIATPSE LEYSGKFAIE DEISFNELPE NYQNRLVLR ILRDNGISDH ALSRTLQKYR KPKPFGDFEV IPEIRSSVIK HGGDFYLVLH LSHQIRSKKT LWELVGRNKD A LRDFLKEH ...String: MLMKVLTNMV KLNQDIIPNE IYLYKIFNKP EDGMNIYKIA YRNHGIVIDP QNRIIATPSE LEYSGKFAIE DEISFNELPE NYQNRLVLR ILRDNGISDH ALSRTLQKYR KPKPFGDFEV IPEIRSSVIK HGGDFYLVLH LSHQIRSKKT LWELVGRNKD A LRDFLKEH RGTILLRDIA SEHKVVYKPI FKRYNGDPDL IEDNSNDVEH WYDYHLERYW NTPELKKEFY KKFGPVDLNQ PI ILAKPLR QHNRGDLVHL LPQFVVPVYN AEQLNDILAS EILEYLKLTS NQRISLLSRL INDIKTNTNI IVSSLTELEA NTF DVDLND MLQVRNADNV KVTLSELEIS KTRLFTWMKS RKYPVILPYD IPQKLKKIEK IPVFIIVDSA LSRDIQTFAK DEFR YLISS LQKSLSNWVD FPILDIRDKY IFTIDLTSDK DIVNLSIKLV NLMKNAELGL ALIATRTKLP NETFDEVKKR LFSVN IISQ VVNEATLYKR DKYNESRLNL YVQHNLLFQI LSKLGIKYYV LRHKFSYDYI VGIDVTPMKL SHGYIGGSAV MFDSQG YIR KIIPVEIGEQ MGESIDMKEF FKDMVVQFGK FGIDLEGKSI LILRDGKITK DEEEGLAYIS KVFGIKITTF NIVKRHL LR IFANRKLYLR LANSVYLLPH RIKQSVGTPV PLKLSEKRLI LDGTITSQEI TYNDIFEILL LSELNYGSIS ADMKLPAP V HYAHKFVRAL RKGWRIREEL LAEGFLYFV UniProtKB: Argonaute family protein |
-Macromolecule #2: Target DNA
| Macromolecule | Name: Target DNA / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Theoretical: 5.051302 KDa |
| Sequence | String: (DA)(DC)(DA)(DA)(DC)(DC)(DT)(DA)(DC)(DT) (DA)(DC)(DC)(DT)(DC)(DC)(DT) |
-Macromolecule #3: Guide DNA
| Macromolecule | Name: Guide DNA / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| Molecular weight | Theoretical: 5.049273 KDa |
| Sequence | String: (DG)(DG)(DA)(DG)(DG)(DT)(DA)(DG)(DT)(DA) (DG)(DG)(DT)(DT)(DG)(DT) |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)